RBP Type
Canonical_RBPs
Diseases
Drug
N.A.
Main interacting RNAs
ncRNA
Moonlighting functions
N.A.
Localizations
Stress granucle
BulkPerturb-seq
N.A.
Ensembl ID ENSG00000134627 Gene ID
143689 Accession
18444
Symbol
PIWIL4
Alias
HIWI2;MIWI2
Full Name
piwi like RNA-mediated gene silencing 4
Status
Confidence
Length
77582 bases
Strand
Plus strand
Position
11 : 94543840 - 94621421
RNA binding domain
Piwi , PAZ
Summary
PIWIL4 belongs to the Argonaute family of proteins, which function in development and maintenance of germline stem cells (Sasaki et al., 2003 [PubMed 12906857]).[supplied by OMIM, Mar 2008]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000299001
Piwi
PF02171 4e-88
1
ENSP00000299001
PAZ
PF02170 2.2e-31
2
ENSP00000439710
Piwi
PF02171 2.7e-56
1
ENSP00000444575
PAZ
PF02170 8.4e-32
1
ENSP00000413838
PAZ
PF02170 9.9e-32
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
32267490 Small nucleolar RNAs: continuing identification of novel members and increasing diversity of their molecular mechanisms of action.
Danny Bergeron
2020-04-29
Biochemical Society transactions
36930678 PCB: A pseudotemporal causality-based Bayesian approach to identify EMT-associated regulatory relationships of AS events and RBPs during breast cancer progression.
Liangjie Sun
2023-03-01
PLoS computational biology
25779046 An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes.
Xin He
2015-04-20
Nucleic acids research
37146239 A noncanonical enzymatic function of PIWIL4 maintains genomic integrity and leukemic growth in AML.
Shiva Bamezai
2023-07-06
Blood
31019967 A Simple Competing Endogenous RNA Network Identifies Novel mRNA, miRNA, and lncRNA Markers in Human Cholangiocarcinoma.
Cheng Zhang
2019-01-01
BioMed research international
34876138 PIWIL4 and SUPT5H combine to predict prognosis and immune landscape in intrahepatic cholangiocarcinoma.
Wenbo Zou
2021-12-07
Cancer cell international
Name
Transcript ID
bp
Protein
Translation ID
PIWIL4-204
ENST00000543336
1901
453aa
ENSP00000444575
PIWIL4-202
ENST00000446230
3759
495aa
ENSP00000413838
PIWIL4-201
ENST00000299001
3139
852aa
ENSP00000299001
PIWIL4-205
ENST00000545603
587
118aa
ENSP00000440499
PIWIL4-203
ENST00000537419
831
203aa
ENSP00000439710
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
17596 Alcoholism 8.05812e-005 22978509 54338 Fructose-Bisphosphate Aldolase 6.7720000E-005 - 76088 Attention Deficit Disorder with Hyperactivity 9E-7 18846501 84256 Prion Diseases 4E-6 25897833 84257 Mood Disorders 4E-6 25897833 103077 Prion Diseases 8E-6 25897833 103078 Mood Disorders 8E-6 25897833
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
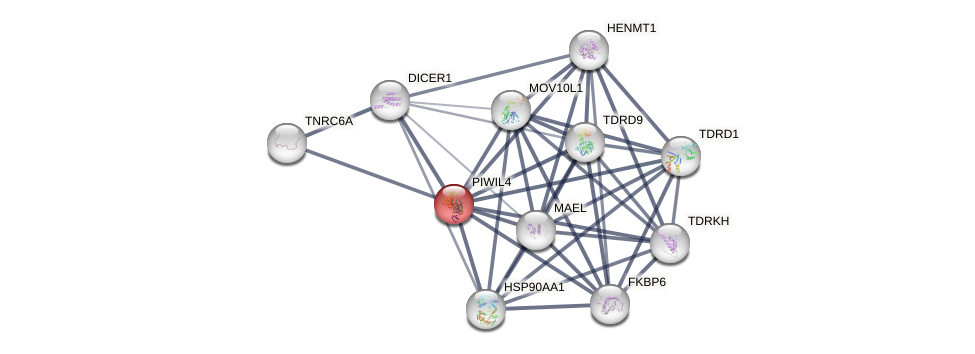
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000134627 homo_sapiens ENSP00000479524 57.3883 38.9462 homo_sapiens ENSP00000299001 58.8028 39.9061 ENSG00000134627 homo_sapiens ENSP00000245255 67.1312 49.1289 homo_sapiens ENSP00000299001 67.8404 49.6479 ENSG00000134627 homo_sapiens ENSP00000349208 49.2292 31.963 homo_sapiens ENSP00000299001 56.2207 36.5023
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000134627 homo_sapiens ENSP00000299001 94.4836 94.0141 nomascus_leucogenys ENSNLEP00000008674 99.2602 98.767 ENSG00000134627 homo_sapiens ENSP00000299001 100 99.5305 pan_paniscus ENSPPAP00000016200 100 99.5305 ENSG00000134627 homo_sapiens ENSP00000299001 99.1784 98.5916 pongo_abelii ENSPPYP00000004327 99.1784 98.5916 ENSG00000134627 homo_sapiens ENSP00000299001 99.8826 99.4131 pan_troglodytes ENSPTRP00000087625 99.8826 99.4131 ENSG00000134627 homo_sapiens ENSP00000299001 99.7653 99.1784 gorilla_gorilla ENSGGOP00000015886 99.7653 99.1784 ENSG00000134627 homo_sapiens ENSP00000299001 82.6291 69.4836 sarcophilus_harrisii ENSSHAP00000020539 81.0127 68.1243 ENSG00000134627 homo_sapiens ENSP00000299001 63.615 54.5775 notamacropus_eugenii ENSMEUP00000004687 68.0905 58.4171 ENSG00000134627 homo_sapiens ENSP00000299001 67.4883 61.5023 choloepus_hoffmanni ENSCHOP00000007116 67.8066 61.7925 ENSG00000134627 homo_sapiens ENSP00000299001 80.6338 71.1268 dasypus_novemcinctus ENSDNOP00000013026 89.3368 78.8036 ENSG00000134627 homo_sapiens ENSP00000299001 46.5962 38.615 erinaceus_europaeus ENSEEUP00000004603 52.5132 43.5185 ENSG00000134627 homo_sapiens ENSP00000299001 47.6526 42.2535 echinops_telfairi ENSETEP00000000411 58.5859 51.9481 ENSG00000134627 homo_sapiens ENSP00000299001 96.0094 92.4883 callithrix_jacchus ENSCJAP00000007483 95.8968 92.3798 ENSG00000134627 homo_sapiens ENSP00000299001 99.2958 98.0047 cercocebus_atys ENSCATP00000012225 99.2958 98.0047 ENSG00000134627 homo_sapiens ENSP00000299001 99.2958 97.5352 macaca_fascicularis ENSMFAP00000030974 99.2958 97.5352 ENSG00000134627 homo_sapiens ENSP00000299001 99.2958 97.5352 macaca_mulatta ENSMMUP00000011747 99.2958 97.5352 ENSG00000134627 homo_sapiens ENSP00000299001 99.2958 97.6526 macaca_nemestrina ENSMNEP00000028109 99.2958 97.6526 ENSG00000134627 homo_sapiens ENSP00000299001 99.1784 97.77 papio_anubis ENSPANP00000009641 99.1784 97.77 ENSG00000134627 homo_sapiens ENSP00000299001 99.061 97.5352 mandrillus_leucophaeus ENSMLEP00000011166 99.061 97.5352 ENSG00000134627 homo_sapiens ENSP00000299001 91.4319 82.1596 tursiops_truncatus ENSTTRP00000010011 91.6471 82.3529 ENSG00000134627 homo_sapiens ENSP00000299001 88.615 79.8122 vulpes_vulpes ENSVVUP00000015790 91.6262 82.5243 ENSG00000134627 homo_sapiens ENSP00000299001 79.108 69.2488 mus_spretus MGP_SPRETEiJ_P0090292 76.7654 67.1982 ENSG00000134627 homo_sapiens ENSP00000299001 92.1361 84.1549 equus_asinus ENSEASP00005061390 92.1361 84.1549 ENSG00000134627 homo_sapiens ENSP00000299001 89.0845 81.1033 capra_hircus ENSCHIP00000004055 88.8759 80.9133 ENSG00000134627 homo_sapiens ENSP00000299001 91.1972 82.277 loxodonta_africana ENSLAFP00000007066 89.8266 81.0405 ENSG00000134627 homo_sapiens ENSP00000299001 90.2582 82.5117 panthera_leo ENSPLOP00000024927 91.1137 83.2938 ENSG00000134627 homo_sapiens ENSP00000299001 89.9061 81.2207 ailuropoda_melanoleuca ENSAMEP00000008013 89.486 80.8411 ENSG00000134627 homo_sapiens ENSP00000299001 83.216 76.1737 bos_indicus_hybrid ENSBIXP00005014217 91.8394 84.0674 ENSG00000134627 homo_sapiens ENSP00000299001 91.1972 82.9812 oryctolagus_cuniculus ENSOCUP00000002484 90.8772 82.6901 ENSG00000134627 homo_sapiens ENSP00000299001 88.8498 80.7512 equus_caballus ENSECAP00000043256 82.1933 74.7014 ENSG00000134627 homo_sapiens ENSP00000299001 90.7277 81.8075 canis_lupus_familiaris ENSCAFP00845027521 90.7277 81.8075 ENSG00000134627 homo_sapiens ENSP00000299001 90.3756 82.5117 panthera_pardus ENSPPRP00000014558 91.2322 83.2938 ENSG00000134627 homo_sapiens ENSP00000299001 90.2582 82.277 marmota_marmota_marmota ENSMMMP00000004149 90.5771 82.5677 ENSG00000134627 homo_sapiens ENSP00000299001 21.7136 19.8357 balaenoptera_musculus ENSBMSP00010010966 91.133 83.2512 ENSG00000134627 homo_sapiens ENSP00000299001 74.8826 67.1361 cavia_porcellus ENSCPOP00000000061 87.8788 78.7879 ENSG00000134627 homo_sapiens ENSP00000299001 90.0235 81.5728 felis_catus ENSFCAP00000010138 90.8768 82.346 ENSG00000134627 homo_sapiens ENSP00000299001 86.7371 78.169 ursus_americanus ENSUAMP00000027338 90.5637 81.6176 ENSG00000134627 homo_sapiens ENSP00000299001 83.4507 70.6573 monodelphis_domestica ENSMODP00000000258 83.3529 70.5744 ENSG00000134627 homo_sapiens ENSP00000299001 88.9671 80.8685 bison_bison_bison ENSBBBP00000012907 89.4923 81.3459 ENSG00000134627 homo_sapiens ENSP00000299001 91.5493 82.1596 monodon_monoceros ENSMMNP00015027131 91.5493 82.1596 ENSG00000134627 homo_sapiens ENSP00000299001 90.3756 81.338 sus_scrofa ENSSSCP00000068835 90.2696 81.2427 ENSG00000134627 homo_sapiens ENSP00000299001 93.662 86.8545 microcebus_murinus ENSMICP00000005822 93.772 86.9565 ENSG00000134627 homo_sapiens ENSP00000299001 86.1502 77.4648 octodon_degus ENSODEP00000017648 88.0096 79.1367 ENSG00000134627 homo_sapiens ENSP00000299001 88.8498 81.1033 ovis_aries_rambouillet ENSOARP00020031373 88.9542 81.1986 ENSG00000134627 homo_sapiens ENSP00000299001 90.6103 81.9249 ursus_maritimus ENSUMAP00000009453 91.0377 82.3113 ENSG00000134627 homo_sapiens ENSP00000299001 80.6338 71.9484 ochotona_princeps ENSOPRP00000003928 81.3018 72.5444 ENSG00000134627 homo_sapiens ENSP00000299001 86.8545 78.169 delphinapterus_leucas ENSDLEP00000011775 92.1544 82.939 ENSG00000134627 homo_sapiens ENSP00000299001 91.9014 83.8028 physeter_catodon ENSPCTP00005017338 91.9014 83.8028 ENSG00000134627 homo_sapiens ENSP00000299001 79.6948 69.6009 mus_caroli MGP_CAROLIEiJ_P0086748 77.3349 67.5399 ENSG00000134627 homo_sapiens ENSP00000299001 87.9108 77.4648 mus_musculus ENSMUSP00000111308 82.2173 72.4479 ENSG00000134627 homo_sapiens ENSP00000299001 19.8357 17.9577 dipodomys_ordii ENSDORP00000018092 90.8602 82.2581 ENSG00000134627 homo_sapiens ENSP00000299001 84.7418 75.5869 mustela_putorius_furo ENSMPUP00000012159 90.25 80.5 ENSG00000134627 homo_sapiens ENSP00000299001 92.2535 88.7324 aotus_nancymaae ENSANAP00000026866 96.5602 92.8747 ENSG00000134627 homo_sapiens ENSP00000299001 96.5962 92.8404 saimiri_boliviensis_boliviensis ENSSBOP00000015626 96.483 92.7315 ENSG00000134627 homo_sapiens ENSP00000299001 82.9812 74.5305 vicugna_pacos ENSVPAP00000005423 83.0787 74.6181 ENSG00000134627 homo_sapiens ENSP00000299001 98.7089 97.3005 chlorocebus_sabaeus ENSCSAP00000014722 98.7089 97.3005 ENSG00000134627 homo_sapiens ENSP00000299001 76.7606 70.5399 tupaia_belangeri ENSTBEP00000011587 76.4019 70.2103 ENSG00000134627 homo_sapiens ENSP00000299001 85.7981 75.5869 mesocricetus_auratus ENSMAUP00000010604 86.7141 76.3938 ENSG00000134627 homo_sapiens ENSP00000299001 91.4319 81.9249 phocoena_sinus ENSPSNP00000017257 91.4319 81.9249 ENSG00000134627 homo_sapiens ENSP00000299001 91.3146 82.6291 camelus_dromedarius ENSCDRP00005022550 91.4219 82.7262 ENSG00000134627 homo_sapiens ENSP00000299001 81.2207 72.5352 carlito_syrichta ENSTSYP00000005115 91.2929 81.5303 ENSG00000134627 homo_sapiens ENSP00000299001 88.7324 80.9859 panthera_tigris_altaica ENSPTIP00000025026 91.1942 83.2328 ENSG00000134627 homo_sapiens ENSP00000299001 85.6807 76.1737 rattus_norvegicus ENSRNOP00000039460 87.7404 78.0048 ENSG00000134627 homo_sapiens ENSP00000299001 93.4272 87.0892 prolemur_simus ENSPSMP00000030537 93.537 87.1915 ENSG00000134627 homo_sapiens ENSP00000299001 86.5023 73.8263 microtus_ochrogaster ENSMOCP00000023605 86.808 74.0872 ENSG00000134627 homo_sapiens ENSP00000299001 51.4085 45.0704 sorex_araneus ENSSARP00000008919 56.0102 49.1049 ENSG00000134627 homo_sapiens ENSP00000299001 86.0329 75.2347 peromyscus_maniculatus_bairdii ENSPEMP00000028291 86.3369 75.5006 ENSG00000134627 homo_sapiens ENSP00000299001 89.2019 81.1033 bos_mutus ENSBMUP00000012341 89.3067 81.1986 ENSG00000134627 homo_sapiens ENSP00000299001 79.2253 69.6009 mus_spicilegus ENSMSIP00000019822 76.9669 67.6169 ENSG00000134627 homo_sapiens ENSP00000299001 85.7981 75.5869 cricetulus_griseus_chok1gshd ENSCGRP00001017995 87.5449 77.1257 ENSG00000134627 homo_sapiens ENSP00000299001 84.9765 78.169 pteropus_vampyrus ENSPVAP00000006837 84.5794 77.8037 ENSG00000134627 homo_sapiens ENSP00000299001 90.8451 82.7465 myotis_lucifugus ENSMLUP00000001838 90.9518 82.8437 ENSG00000134627 homo_sapiens ENSP00000299001 84.507 72.0657 vombatus_ursinus ENSVURP00010013844 84.507 72.0657 ENSG00000134627 homo_sapiens ENSP00000299001 90.7277 84.1549 rhinolophus_ferrumequinum ENSRFEP00010031109 86.3687 80.1117 ENSG00000134627 homo_sapiens ENSP00000299001 90.1408 80.1643 neovison_vison ENSNVIP00000007029 90.1408 80.1643 ENSG00000134627 homo_sapiens ENSP00000299001 87.9108 79.8122 bos_taurus ENSBTAP00000019851 86.7903 78.7949 ENSG00000134627 homo_sapiens ENSP00000299001 90.3756 82.1596 heterocephalus_glaber_female ENSHGLP00000040355 90.1639 81.9672 ENSG00000134627 homo_sapiens ENSP00000299001 20.5399 18.662 heterocephalus_glaber_female ENSHGLP00000057371 87.5 79.5 ENSG00000134627 homo_sapiens ENSP00000299001 98.5916 97.3005 rhinopithecus_bieti ENSRBIP00000031870 98.5916 97.3005 ENSG00000134627 homo_sapiens ENSP00000299001 89.0845 80.3991 canis_lupus_dingo ENSCAFP00020020884 90.3571 81.5476 ENSG00000134627 homo_sapiens ENSP00000299001 90.6103 82.7465 sciurus_vulgaris ENSSVLP00005012190 90.7168 82.8437 ENSG00000134627 homo_sapiens ENSP00000299001 83.9202 71.2441 phascolarctos_cinereus ENSPCIP00000041941 83.9202 71.2441 ENSG00000134627 homo_sapiens ENSP00000299001 87.7934 79.4601 nannospalax_galili ENSNGAP00000023875 88 79.6471 ENSG00000134627 homo_sapiens ENSP00000299001 89.0845 81.2207 bos_grunniens ENSBGRP00000016890 89.2941 81.4118 ENSG00000134627 homo_sapiens ENSP00000299001 48.5915 44.2488 chinchilla_lanigera ENSCLAP00000000448 90.5908 82.4945 ENSG00000134627 homo_sapiens ENSP00000299001 89.2019 81.1033 cervus_hanglu_yarkandensis ENSCHYP00000003740 89.2019 81.1033 ENSG00000134627 homo_sapiens ENSP00000299001 90.7277 80.3991 catagonus_wagneri ENSCWAP00000002422 90.6213 80.3048 ENSG00000134627 homo_sapiens ENSP00000299001 49.7653 45.7746 ictidomys_tridecemlineatus ENSSTOP00000006211 92.9825 85.5263 ENSG00000134627 homo_sapiens ENSP00000299001 92.723 85.446 otolemur_garnettii ENSOGAP00000010876 92.0746 84.8485 ENSG00000134627 homo_sapiens ENSP00000299001 96.7136 93.5446 cebus_imitator ENSCCAP00000003169 96.6002 93.4349 ENSG00000134627 homo_sapiens ENSP00000299001 90.0235 81.8075 urocitellus_parryii ENSUPAP00010002391 89.8126 81.6159 ENSG00000134627 homo_sapiens ENSP00000299001 89.0845 80.1643 moschus_moschiferus ENSMMSP00000022871 89.0845 80.1643 ENSG00000134627 homo_sapiens ENSP00000299001 98.8263 97.6526 rhinopithecus_roxellana ENSRROP00000003777 98.8263 97.6526 ENSG00000134627 homo_sapiens ENSP00000299001 88.0282 77.3474 mus_pahari MGP_PahariEiJ_P0022408 88.4434 77.7123 ENSG00000134627 homo_sapiens ENSP00000299001 94.3662 87.9108 propithecus_coquereli ENSPCOP00000020573 94.4771 88.0141 ENSG00000134627 homo_sapiens ENSP00000299001 49.8826 30.3991 caenorhabditis_elegans D2030.6.1 51.5777 31.432 ENSG00000134627 homo_sapiens ENSP00000299001 50.939 32.6291 drosophila_melanogaster FBpp0079755 51.4828 32.9775 ENSG00000134627 homo_sapiens ENSP00000299001 52.1127 32.0423 drosophila_melanogaster FBpp0079754 51.2702 31.5243 ENSG00000134627 homo_sapiens ENSP00000299001 64.6714 45.4225 petromyzon_marinus ENSPMAP00000010233 63.6994 44.7399 ENSG00000134627 homo_sapiens ENSP00000299001 64.4366 43.1925 eptatretus_burgeri ENSEBUP00000005507 63.3218 42.4452 ENSG00000134627 homo_sapiens ENSP00000299001 69.1315 54.8122 callorhinchus_milii ENSCMIP00000029465 72.4477 57.4416 ENSG00000134627 homo_sapiens ENSP00000299001 74.1784 57.7465 xenopus_tropicalis ENSXETP00000014018 74.0914 57.6788 ENSG00000134627 homo_sapiens ENSP00000299001 76.9953 62.9108 chrysemys_picta_bellii ENSCPBP00000034460 84.8642 69.3402 ENSG00000134627 homo_sapiens ENSP00000299001 78.2864 63.3803 sphenodon_punctatus ENSSPUP00000005645 75.1973 60.8794 ENSG00000134627 homo_sapiens ENSP00000299001 76.9953 62.4413 crocodylus_porosus ENSCPRP00005021891 83.4606 67.6845 ENSG00000134627 homo_sapiens ENSP00000299001 66.3146 52.23 laticauda_laticaudata ENSLLTP00000024761 76.7663 60.462 ENSG00000134627 homo_sapiens ENSP00000299001 72.3005 56.9249 notechis_scutatus ENSNSUP00000007264 79.6895 62.7426 ENSG00000134627 homo_sapiens ENSP00000299001 73.4742 57.8638 pseudonaja_textilis ENSPTXP00000006541 78.8413 62.0907 ENSG00000134627 homo_sapiens ENSP00000299001 75.3521 57.6291 anolis_carolinensis ENSACAP00000013820 75.8865 58.0378 ENSG00000134627 homo_sapiens ENSP00000299001 69.953 54.5775 naja_naja ENSNNAP00000005866 77.7053 60.6258 ENSG00000134627 homo_sapiens ENSP00000299001 75.4695 59.6244 podarcis_muralis ENSPMRP00000012958 77.4699 61.2048 ENSG00000134627 homo_sapiens ENSP00000299001 18.662 14.3192 podarcis_muralis ENSPMRP00000035055 78.7129 60.396 ENSG00000134627 homo_sapiens ENSP00000299001 77.8169 61.385 salvator_merianae ENSSMRP00000013065 80.2663 63.3172 ENSG00000134627 homo_sapiens ENSP00000299001 82.0423 66.6667 pelodiscus_sinensis ENSPSIP00000015955 81.6589 66.3551 ENSG00000134627 homo_sapiens ENSP00000299001 80.6338 66.0798 gopherus_evgoodei ENSGEVP00005010069 81.2057 66.5485 ENSG00000134627 homo_sapiens ENSP00000299001 82.3944 66.9014 chelonoidis_abingdonii ENSCABP00000030826 84.1727 68.3453 ENSG00000134627 homo_sapiens ENSP00000299001 83.8028 68.5446 terrapene_carolina_triunguis ENSTMTP00000028516 83.0233 67.907 ENSG00000134627 homo_sapiens ENSP00000299001 55.9859 38.7324 ciona_savignyi ENSCSAVP00000005362 60.8418 42.0918 ENSG00000134627 homo_sapiens ENSP00000299001 71.0094 55.2817 leptobrachium_leishanense ENSLLEP00000001527 75.5306 58.8015
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0004521 enables RNA endonuclease activity 2 IBA,ISS Homo_sapiens(9606) Function GO:0005634 located_in nucleus 2 IDA,ISS Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 1 IDA Homo_sapiens(9606) Component GO:0005737 located_in cytoplasm 2 IDA,ISS Homo_sapiens(9606) Component GO:0005739 located_in mitochondrion 1 IDA Homo_sapiens(9606) Component GO:0006417 involved_in regulation of translation 1 IEA Homo_sapiens(9606) Process GO:0007283 involved_in spermatogenesis 1 ISS Homo_sapiens(9606) Process GO:0010669 involved_in epithelial structure maintenance 1 IMP Homo_sapiens(9606) Process GO:0031047 involved_in regulatory ncRNA-mediated gene silencing 2 IBA,ISS Homo_sapiens(9606) Process GO:0034584 enables piRNA binding 3 IBA,IDA,ISS Homo_sapiens(9606) Function GO:0043186 is_active_in P granule 2 IBA,ISS Homo_sapiens(9606) Component GO:0051321 involved_in meiotic cell cycle 1 IEA Homo_sapiens(9606) Process GO:0071514 involved_in genomic imprinting 1 ISS Homo_sapiens(9606) Process GO:0071547 located_in piP-body 1 ISS Homo_sapiens(9606) Component GO:0140965 involved_in secondary piRNA processing 1 ISS Homo_sapiens(9606) Process GO:0141005 acts_upstream_of_positive_effect retrotransposon silencing by heterochromatin formation 1 ISS Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.