RBP Type
Canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
E3 ligase
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000132274 Gene ID
10346 Accession
16379
Symbol
TRIM22
Alias
RNF94;STAF50;GPSTAF50
Full Name
tripartite motif containing 22
Status
Confidence
Length
47393 bases
Strand
Plus strand
Position
11 : 5689697 - 5737089
RNA binding domain
SPRY
Summary
The protein encoded by this gene is a member of the tripartite motif (TRIM) family. The TRIM motif includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. This protein localizes to the cytoplasm and its expression is induced by interferon. The protein is involved in innate immunity against different DNA and RNA viruses. [provided by RefSeq, Oct 2021]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000514104
SPRY
PF00622 1.1e-07
1
ENSP00000393250
SPRY
PF00622 1.6e-07
1
ENSP00000369299
SPRY
PF00622 1.8e-07
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
19624813 Prediction of acute multiple sclerosis relapses by transcription levels of peripheral blood cells.
Michael Gurevich
2009-07-22
BMC medical genomics
Name
Transcript ID
bp
Protein
Translation ID
TRIM22-209
ENST00000460454
4528
No protein
-
TRIM22-212
ENST00000699052
2708
No protein
-
TRIM22-204
ENST00000425490
812
203aa
ENSP00000400417
TRIM22-213
ENST00000699053
2062
277aa
ENSP00000514103
TRIM22-214
ENST00000699054
4185
No protein
-
TRIM22-210
ENST00000480395
3750
No protein
-
TRIM22-201
ENST00000379965
2888
498aa
ENSP00000369299
TRIM22-215
ENST00000699055
3355
No protein
-
TRIM22-216
ENST00000699056
5084
No protein
-
TRIM22-208
ENST00000454828
2684
466aa
ENSP00000393250
TRIM22-202
ENST00000414641
897
227aa
ENSP00000396849
TRIM22-217
ENST00000699057
1699
387aa
ENSP00000514104
TRIM22-203
ENST00000414897
655
143aa
ENSP00000388994
TRIM22-218
ENST00000699058
5395
No protein
-
TRIM22-211
ENST00000493494
460
No protein
-
TRIM22-207
ENST00000450670
508
63aa
ENSP00000406412
TRIM22-205
ENST00000429063
1241
37aa
ENSP00000409991
TRIM22-206
ENST00000444844
1244
38aa
ENSP00000404204
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
26830 Blood Pressure 9.3910000E-005 - 41772 Platelet Function Tests 1.1102600E-005 -
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
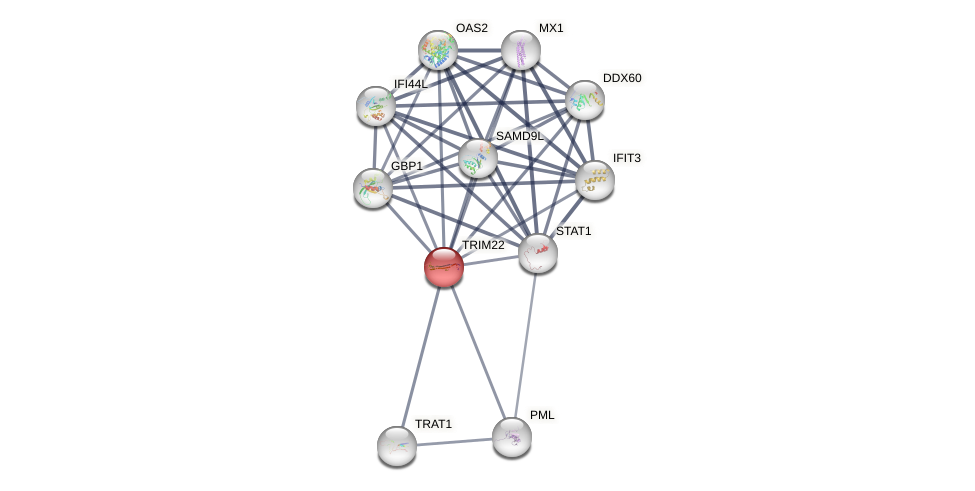
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000209 involved_in protein polyubiquitination 1 IBA Homo_sapiens(9606) Process GO:0002230 involved_in positive regulation of defense response to virus by host 1 IDA Homo_sapiens(9606) Process GO:0003713 enables transcription coactivator activity 1 IDA Homo_sapiens(9606) Function GO:0003714 enables transcription corepressor activity 1 TAS Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 located_in nucleus 1 IDA Homo_sapiens(9606) Component GO:0005654 is_active_in nucleoplasm 2 IBA,IDA Homo_sapiens(9606) Component GO:0005737 is_active_in cytoplasm 2 IBA,IDA Homo_sapiens(9606) Component GO:0005794 located_in Golgi apparatus 1 IDA Homo_sapiens(9606) Component GO:0005829 is_active_in cytosol 2 IBA,TAS Homo_sapiens(9606) Component GO:0006355 involved_in regulation of DNA-templated transcription 1 TAS Homo_sapiens(9606) Process GO:0006955 involved_in immune response 1 TAS Homo_sapiens(9606) Process GO:0008270 enables zinc ion binding 1 IEA Homo_sapiens(9606) Function GO:0009615 involved_in response to virus 1 TAS Homo_sapiens(9606) Process GO:0010508 involved_in positive regulation of autophagy 2 IBA,IMP Homo_sapiens(9606) Process GO:0015030 located_in Cajal body 1 IEA Homo_sapiens(9606) Component GO:0016604 located_in nuclear body 1 IDA Homo_sapiens(9606) Component GO:0016607 located_in nuclear speck 1 IEA Homo_sapiens(9606) Component GO:0019901 enables protein kinase binding 2 IBA,IPI Homo_sapiens(9606) Function GO:0030674 enables protein-macromolecule adaptor activity 1 IPI Homo_sapiens(9606) Function GO:0032880 involved_in regulation of protein localization 2 IBA,IMP Homo_sapiens(9606) Process GO:0042802 enables identical protein binding 1 IPI Homo_sapiens(9606) Function GO:0042803 enables protein homodimerization activity 1 IBA Homo_sapiens(9606) Function GO:0043123 involved_in positive regulation of I-kappaB kinase/NF-kappaB signaling 2 IBA,IDA Homo_sapiens(9606) Process GO:0045087 involved_in innate immune response 1 IBA Homo_sapiens(9606) Process GO:0045892 involved_in negative regulation of DNA-templated transcription 1 IEA Homo_sapiens(9606) Process GO:0045893 involved_in positive regulation of DNA-templated transcription 1 IEA Homo_sapiens(9606) Process GO:0046596 involved_in regulation of viral entry into host cell 1 IBA Homo_sapiens(9606) Process GO:0051091 involved_in positive regulation of DNA-binding transcription factor activity 1 IDA Homo_sapiens(9606) Process GO:0051092 involved_in positive regulation of NF-kappaB transcription factor activity 2 IBA,IDA Homo_sapiens(9606) Process GO:0051607 involved_in defense response to virus 1 IEA Homo_sapiens(9606) Process GO:0061630 enables ubiquitin protein ligase activity 2 IBA,IDA Homo_sapiens(9606) Function GO:0070534 involved_in protein K63-linked ubiquitination 1 IDA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.