| GO:0001701 | involved_in in utero embryonic development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001750 | located_in photoreceptor outer segment | 1 | IDA | Homo_sapiens(9606) | Component |
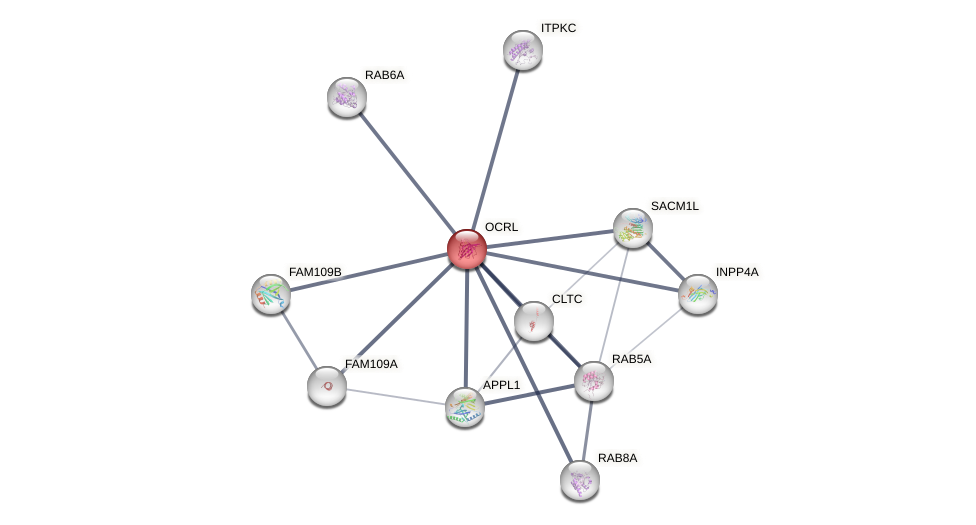
| GO:0004439 | enables phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity | 2 | IBA,TAS | Homo_sapiens(9606) | Function |
| GO:0004445 | enables inositol-polyphosphate 5-phosphatase activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005096 | enables GTPase activator activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005764 | located_in lysosome | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005769 | located_in early endosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005795 | located_in Golgi stack | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005798 | located_in Golgi-associated vesicle | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005802 | located_in trans-Golgi network | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005886 | located_in plasma membrane | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005905 | located_in clathrin-coated pit | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0006629 | involved_in lipid metabolic process | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0006661 | involved_in phosphatidylinositol biosynthetic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007165 | involved_in signal transduction | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0016020 | is_active_in membrane | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0030136 | located_in clathrin-coated vesicle | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0030670 | located_in phagocytic vesicle membrane | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0031267 | enables small GTPase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0031901 | located_in early endosome membrane | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0034485 | enables phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0034596 | enables phosphatidylinositol phosphate 4-phosphatase activity | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0036092 | involved_in phosphatidylinositol-3-phosphate biosynthetic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0043087 | involved_in regulation of GTPase activity | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0043647 | involved_in inositol phosphate metabolic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0043813 | enables phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0046856 | involved_in phosphatidylinositol dephosphorylation | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0052658 | enables inositol-1,4,5-trisphosphate 5-phosphatase activity | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0052659 | enables inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0052745 | enables inositol phosphate phosphatase activity | 2 | IDA,NAS | Homo_sapiens(9606) | Function |
| GO:0060271 | involved_in cilium assembly | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0061024 | involved_in membrane organization | 1 | TAS | Homo_sapiens(9606) | Process |