RBP Type
Non-canonical_RBPs
Diseases
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
Metabolic enzyme
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000119711 Gene ID
4329 Accession
7179
Symbol
ALDH6A1
Alias
MMSDH;MMSADHA
Full Name
aldehyde dehydrogenase 6 family member A1
Status
Confidence
Length
27607 bases
Strand
Minus strand
Position
14 : 74056847 - 74084453
RNA binding domain
N.A.
Summary
This gene encodes a member of the aldehyde dehydrogenase protein family. The encoded protein is a mitochondrial methylmalonate semialdehyde dehydrogenase that plays a role in the valine and pyrimidine catabolic pathways. This protein catalyzes the irreversible oxidative decarboxylation of malonate and methylmalonate semialdehydes to acetyl- and propionyl-CoA. Methylmalonate semialdehyde dehydrogenase deficiency is characterized by elevated beta-alanine, 3-hydroxypropionic acid, and both isomers of 3-amino and 3-hydroxyisobutyric acids in urine organic acids. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Jun 2013]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
31213602 CAPRI enables comparison of evolutionarily conserved RNA interacting regions.
Amol Panhale
2019-06-18
Nature communications
Name
Transcript ID
bp
Protein
Translation ID
ALDH6A1-202
ENST00000553458
5462
535aa
ENSP00000450436
ALDH6A1-205
ENST00000554501
2221
No protein
-
ALDH6A1-201
ENST00000350259
1869
522aa
ENSP00000342564
ALDH6A1-206
ENST00000555126
1382
252aa
ENSP00000452081
ALDH6A1-204
ENST00000554231
1323
No protein
-
ALDH6A1-203
ENST00000553814
580
No protein
-
ALDH6A1-207
ENST00000556852
726
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
44416 Platelet Function Tests 5.4178840E-010 - 44422 Platelet Function Tests 7.8994069E-007 - 44492 Platelet Function Tests 5.4178840E-010 - 44498 Platelet Function Tests 7.8994069E-007 - 96134 Circadian Rhythm 3E-7 20585627
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
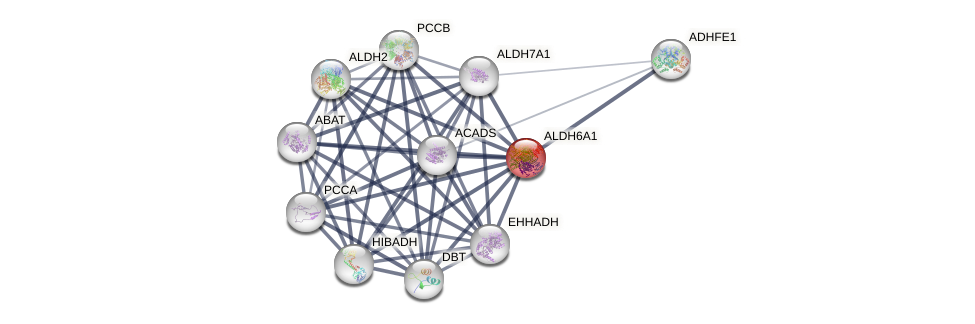
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000062 enables fatty-acyl-CoA binding 1 ISS Homo_sapiens(9606) Function GO:0003723 enables RNA binding 1 HDA Homo_sapiens(9606) Function GO:0004491 enables methylmalonate-semialdehyde dehydrogenase (acylating) activity 4 IBA,IMP,ISS,TAS Homo_sapiens(9606) Function GO:0005654 located_in nucleoplasm 1 IDA Homo_sapiens(9606) Component GO:0005739 is_active_in mitochondrion 3 IBA,IDA,NAS Homo_sapiens(9606) Component GO:0005759 located_in mitochondrial matrix 1 TAS Homo_sapiens(9606) Component GO:0006210 involved_in thymine catabolic process 2 IBA,IMP Homo_sapiens(9606) Process GO:0006573 involved_in valine metabolic process 1 ISS Homo_sapiens(9606) Process GO:0006574 involved_in valine catabolic process 2 IBA,IMP Homo_sapiens(9606) Process GO:0009083 involved_in branched-chain amino acid catabolic process 1 TAS Homo_sapiens(9606) Process GO:0018478 enables malonate-semialdehyde dehydrogenase (acetylating) activity 2 IBA,ISS Homo_sapiens(9606) Function GO:0019859 involved_in thymine metabolic process 1 ISS Homo_sapiens(9606) Process GO:0050873 involved_in brown fat cell differentiation 1 IEA Homo_sapiens(9606) Process GO:0102662 enables malonate-semialdehyde dehydrogenase (acetylating, NAD+) activity 1 IEA Homo_sapiens(9606) Function
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.