Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | CancerSARS-COV-OC43SARS-COV-2 |
| Drug | N.A. |
| Main interacting RNAs | ncRNA |
| Moonlighting functions | N.A. |
| Localizations | N.A. |
| BulkPerturb-seq | DataSet_02_76 |
Description
| Ensembl ID | ENSG00000119705 | Gene ID | 81892 | Accession | 20495 |
| Symbol | SLIRP | Alias | DC50;PD04872;C14orf156 | Full Name | SRA stem-loop interacting RNA binding protein |
| Status | Confidence | Length | 53034 bases | Strand | Plus strand |
| Position | 14 : 77708071 - 77761104 | RNA binding domain | RRM_1 | ||
| Summary | Steroid receptor RNA activator (SRA, or SRA1; MIM 603819) is a complex RNA molecule containing multiple stable stem-loop structures that functions in coactivation of nuclear receptors. SLIRP interacts with stem-loop structure-7 of SRA (STR7) and modulates nuclear receptor transactivation (Hatchell et al., 2006 [PubMed 16762838]).[supplied by OMIM, Mar 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000452329 | RRM_1 | PF00076 | 4.1e-13 | 1 |
| ENSP00000452057 | RRM_1 | PF00076 | 6.2e-13 | 1 |
| ENSP00000478794 | RRM_1 | PF00076 | 6.8e-13 | 1 |
| ENSP00000450770 | RRM_1 | PF00076 | 8.6e-13 | 1 |
| ENSP00000238688 | RRM_1 | PF00076 | 1.1e-12 | 1 |
| ENSP00000450909 | RRM_1 | PF00076 | 1.1e-12 | 1 |
| ENSP00000450849 | RRM_1 | PF00076 | 1.5e-12 | 1 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 28814717 | Drosophila protease ClpXP specifically degrades DmLRPPRC1 controlling mitochondrial mRNA and translation. | Yuichi Matsushima | 2017-08-16 | Scientific reports |
| 22661577 | LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. | Takeshi Chujo | 2012-09-01 | Nucleic acids research |
| 26016801 | Mitochondrial transcript maturation and its disorders. | Lindsey L Van | 2015-07-01 | Journal of inherited metabolic disease |
| 22808186 | Knockdown of the cellular protein LRPPRC attenuates HIV-1 infection. | J Cameron Schweitzer | 2012-01-01 | PloS one |
| 33312877 | Poly (A) tail length of human mitochondrial mRNAs is tissue-specific and a mutation in LRPPRC results in transcript-specific patterns of deadenylation. | Shamisa Honarmand | 2020-12-01 | Molecular genetics and metabolism reports |
| 26866271 | Affinity purification-mass spectrometry analysis of bcl-2 interactome identified SLIRP as a novel interacting protein. | D Trisciuoglio | 2016-02-11 | Cell death & disease |
| 25214534 | Tissue-specific responses to the LRPPRC founder mutation in French Canadian Leigh Syndrome. | Florin Sasarman | 2015-01-15 | Human molecular genetics |
| 31260285 | SLIRP Interacts with Helicases to Facilitate 2'-O-Methylation of rRNA and to Promote Translation. | Lin Li | 2019-07-17 | Journal of the American Chemical Society |
| 20200222 | LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. | Florin Sasarman | 2010-04-15 | Molecular biology of the cell |
| 31819114 | Interaction between androgen receptor and coregulator SLIRP is regulated by Ack1 tyrosine kinase and androgen. | Dinuka D De | 2019-12-09 | Scientific reports |
| 26510951 | LRPPRC mutations cause early-onset multisystem mitochondrial disease outside of the French-Canadian population. | Monika Oláhová | 2015-12-01 | Brain : a journal of neurology |
| 34426662 | Pathogenic SLIRP variants as a novel cause of autosomal recessive mitochondrial encephalomyopathy with complex I and IV deficiency. | Le Guo | 2021-12-01 | European journal of human genetics : EJHG |
| 27353330 | SLIRP stabilizes LRPPRC via an RRM-PPR protein interface. | Henrik Spåhr | 2016-08-19 | Nucleic acids research |
| 31861673 | Mitochondrial Gene Expression and Beyond-Novel Aspects of Cellular Physiology. | V Anna Kotrys | 2019-12-19 | Cells |
| 23635806 | Human pentatricopeptide proteins: only a few and what do they do? | N Robert Lightowlers | 2013-01-01 | RNA biology |
| 19680543 | A computational screen for regulators of oxidative phosphorylation implicates SLIRP in mitochondrial RNA homeostasis. | M Joshua Baughman | 2009-08-01 | PLoS genetics |
| 37355389 | RNA-binding protein complex AMG-1/SLRP-1 mediates germline development and spermatogenesis by maintaining mitochondrial homeostasis in Caenorhabditis elegans. | Peng Wang | 2023-07-15 | Science bulletin |
| 20220140 | Mitochondrial and nuclear genomic responses to loss of LRPPRC expression. | M Vishal Gohil | 2010-04-30 | The Journal of biological chemistry |
| 34975760 | Ovarian Aging: Role of Pituitary-Ovarian Axis Hormones and ncRNAs in Regulating Ovarian Mitochondrial Activity. | Marco Colella | 2021-01-01 | Frontiers in endocrinology |
| 23275553 | Alternative translation initiation augments the human mitochondrial proteome. | Lawrence Kazak | 2013-02-01 | Nucleic acids research |
| 20144953 | Targeting of the cytosolic poly(A) binding protein PABPC1 to mitochondria causes mitochondrial translation inhibition. | Mateusz Wydro | 2010-06-01 | Nucleic acids research |
| 22022283 | The bicoid stability factor controls polyadenylation and expression of specific mitochondrial mRNAs in Drosophila melanogaster. | Ana Bratic | 2011-10-01 | PLoS genetics |
| 20544879 | A compendium of human mitochondrial gene expression machinery with links to disease. | E Timothy Shutt | 2010-06-01 | Environmental and molecular mutagenesis |
| 31363146 | A protein-RNA interaction atlas of the ribosome biogenesis factor AATF. | J W Rainer Kaiser | 2019-07-30 | Scientific reports |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| SLIRP-212 | ENST00000613856 | 420 | 92aa | ENSP00000478794 |
| SLIRP-201 | ENST00000238688 | 394 | 107aa | ENSP00000238688 |
| SLIRP-209 | ENST00000557342 | 376 | 109aa | ENSP00000450909 |
| SLIRP-211 | ENST00000557623 | 865 | 98aa | ENSP00000452057 |
| SLIRP-210 | ENST00000557431 | 725 | 124aa | ENSP00000450849 |
| SLIRP-204 | ENST00000555890 | 471 | 40aa | ENSP00000451606 |
| SLIRP-207 | ENST00000556831 | 729 | 96aa | ENSP00000450770 |
| SLIRP-203 | ENST00000554074 | 445 | No protein | - |
| SLIRP-205 | ENST00000556310 | 626 | 76aa | ENSP00000452329 |
| SLIRP-206 | ENST00000556375 | 408 | 72aa | ENSP00000452162 |
| SLIRP-202 | ENST00000553981 | 285 | 56aa | ENSP00000450587 |
| SLIRP-208 | ENST00000556956 | 450 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
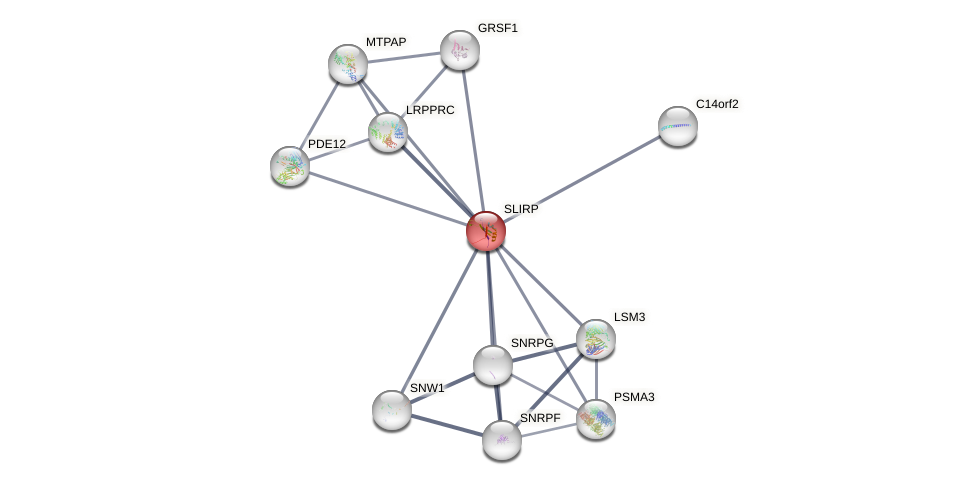
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000119705 | ||||||||
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 84.4037 | 77.9817 | nomascus_leucogenys | ENSNLEP00000032634 | 89.3204 | 82.5243 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 85.3211 | 83.4862 | pan_paniscus | ENSPPAP00000035866 | 75 | 73.3871 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 82.5688 | 75.2294 | pongo_abelii | ENSPPYP00000044169 | 73.1707 | 66.6667 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 85.3211 | 83.4862 | pan_troglodytes | ENSPTRP00000086876 | 75 | 73.3871 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 85.3211 | 83.4862 | gorilla_gorilla | ENSGGOP00000039289 | 75 | 73.3871 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 65.1376 | 48.6239 | ornithorhynchus_anatinus | ENSOANP00000042660 | 65.1376 | 48.6239 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 62.3853 | 48.6239 | sarcophilus_harrisii | ENSSHAP00000041145 | 66.0194 | 51.4563 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 77.0642 | 65.1376 | dasypus_novemcinctus | ENSDNOP00000005980 | 76.3636 | 64.5455 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 69.7248 | 58.7156 | erinaceus_europaeus | ENSEEUP00000006612 | 75.2475 | 63.3663 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 84.4037 | 66.9725 | echinops_telfairi | ENSETEP00000008402 | 85.1852 | 67.5926 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 79.8165 | 68.8073 | callithrix_jacchus | ENSCJAP00000052809 | 70.1613 | 60.4839 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 80.7339 | 71.5596 | cercocebus_atys | ENSCATP00000025739 | 78.5714 | 69.6429 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 82.5688 | 72.4771 | macaca_fascicularis | ENSMFAP00000022503 | 80.3571 | 70.5357 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 92.6606 | 81.6514 | macaca_mulatta | ENSMMUP00000032696 | 92.6606 | 81.6514 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 81.6514 | 72.4771 | macaca_nemestrina | ENSMNEP00000008888 | 79.4643 | 70.5357 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 81.6514 | 71.5596 | papio_anubis | ENSPANP00000027382 | 79.4643 | 69.6429 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 80.7339 | 71.5596 | mandrillus_leucophaeus | ENSMLEP00000039211 | 78.5714 | 69.6429 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 80.7339 | 68.8073 | vulpes_vulpes | ENSVVUP00000005676 | 67.1756 | 57.2519 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 83.4862 | 66.9725 | mus_spretus | MGP_SPRETEiJ_P0034518 | 81.25 | 65.1786 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 78.8991 | 66.055 | equus_asinus | ENSEASP00005041150 | 87.7551 | 73.4694 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 78.8991 | 65.1376 | bos_indicus_hybrid | ENSBIXP00005040444 | 73.5043 | 60.6838 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 83.4862 | 67.8899 | equus_caballus | ENSECAP00000048493 | 80.531 | 65.4867 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 80.7339 | 69.7248 | canis_lupus_familiaris | ENSCAFP00845010564 | 67.6923 | 58.4615 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 84.4037 | 71.5596 | canis_lupus_familiaris | ENSCAFP00845040275 | 82.8829 | 70.2703 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 77.9817 | 66.055 | panthera_pardus | ENSPPRP00000024127 | 86.7347 | 73.4694 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 87.156 | 69.7248 | marmota_marmota_marmota | ENSMMMP00000022180 | 85.5856 | 68.4685 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 83.4862 | 68.8073 | balaenoptera_musculus | ENSBMSP00010011340 | 84.2593 | 69.4444 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 89.9083 | 74.3119 | felis_catus | ENSFCAP00000052200 | 88.2883 | 72.973 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 87.156 | 74.3119 | ursus_americanus | ENSUAMP00000029116 | 85.5856 | 72.973 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 78.8991 | 64.2202 | bison_bison_bison | ENSBBBP00000012951 | 73.5043 | 59.8291 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 87.156 | 71.5596 | monodon_monoceros | ENSMMNP00015019329 | 85.5856 | 70.2703 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 86.2385 | 67.8899 | sus_scrofa | ENSSSCP00000066201 | 63.9456 | 50.3401 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 77.0642 | 66.055 | microcebus_murinus | ENSMICP00000050038 | 71.7949 | 61.5385 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 86.2385 | 71.5596 | octodon_degus | ENSODEP00000017077 | 83.9286 | 69.6429 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 78.8991 | 66.055 | ovis_aries_rambouillet | ENSOARP00020006383 | 73.5043 | 61.5385 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 88.0734 | 74.3119 | ursus_maritimus | ENSUMAP00000009485 | 86.4865 | 72.973 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 86.2385 | 70.6422 | delphinapterus_leucas | ENSDLEP00000019864 | 84.6847 | 69.3694 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 81.6514 | 66.055 | mus_caroli | MGP_CAROLIEiJ_P0032999 | 82.4074 | 66.6667 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 83.4862 | 66.9725 | mus_musculus | ENSMUSP00000125341 | 81.25 | 65.1786 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 86.2385 | 66.9725 | dipodomys_ordii | ENSDORP00000011639 | 84.6847 | 65.7658 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 89.9083 | 73.3945 | mustela_putorius_furo | ENSMPUP00000008666 | 36.9811 | 30.1887 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 77.0642 | 66.9725 | saimiri_boliviensis_boliviensis | ENSSBOP00000006134 | 67.2 | 58.4 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 83.4862 | 67.8899 | vicugna_pacos | ENSVPAP00000001644 | 84.2593 | 68.5185 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 93.578 | 82.5688 | chlorocebus_sabaeus | ENSCSAP00000007939 | 93.578 | 82.5688 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 81.6514 | 66.9725 | mesocricetus_auratus | ENSMAUP00000003475 | 80.1802 | 65.7658 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 87.156 | 71.5596 | phocoena_sinus | ENSPSNP00000022683 | 85.5856 | 70.2703 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 44.9541 | 38.5321 | carlito_syrichta | ENSTSYP00000027205 | 87.5 | 75 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 77.9817 | 66.055 | panthera_tigris_altaica | ENSPTIP00000014943 | 86.7347 | 73.4694 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 68.8073 | 53.211 | rattus_norvegicus | ENSRNOP00000080791 | 73.5294 | 56.8627 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 76.1468 | 64.2202 | prolemur_simus | ENSPSMP00000006524 | 55.7047 | 46.9799 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 81.6514 | 64.2202 | microtus_ochrogaster | ENSMOCP00000017416 | 79.4643 | 62.5 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 79.8165 | 59.633 | peromyscus_maniculatus_bairdii | ENSPEMP00000013097 | 76.3158 | 57.0175 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 77.9817 | 64.2202 | bos_mutus | ENSBMUP00000024276 | 72.6496 | 59.8291 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 83.4862 | 66.9725 | mus_spicilegus | ENSMSIP00000014762 | 81.25 | 65.1786 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 81.6514 | 65.1376 | cricetulus_griseus_chok1gshd | ENSCGRP00001026884 | 80.1802 | 63.964 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 82.5688 | 65.1376 | myotis_lucifugus | ENSMLUP00000022109 | 81.8182 | 64.5455 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 85.3211 | 66.9725 | myotis_lucifugus | ENSMLUP00000016574 | 83.7838 | 65.7658 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 83.4862 | 66.9725 | rhinolophus_ferrumequinum | ENSRFEP00010024676 | 82.7273 | 66.3636 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 74.3119 | 60.5505 | rhinolophus_ferrumequinum | ENSRFEP00010021526 | 35.8407 | 29.2035 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 90.8257 | 74.3119 | neovison_vison | ENSNVIP00000017922 | 48.2927 | 39.5122 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 80.7339 | 60.5505 | heterocephalus_glaber_female | ENSHGLP00000035183 | 79.2793 | 59.4595 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 77.9817 | 55.9633 | heterocephalus_glaber_female | ENSHGLP00000049149 | 75.8929 | 54.4643 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 81.6514 | 60.5505 | heterocephalus_glaber_female | ENSHGLP00000030363 | 80.1802 | 59.4595 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 72.4771 | 56.8807 | heterocephalus_glaber_female | ENSHGLP00000031599 | 78.2178 | 61.3861 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 69.7248 | 55.0459 | heterocephalus_glaber_female | ENSHGLP00000058626 | 81.7204 | 64.5161 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 80.7339 | 60.5505 | heterocephalus_glaber_female | ENSHGLP00000034388 | 79.2793 | 59.4595 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 76.1468 | 54.1284 | heterocephalus_glaber_female | ENSHGLP00000060431 | 72.1739 | 51.3043 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 67.8899 | 49.5413 | heterocephalus_glaber_female | ENSHGLP00000043996 | 82.2222 | 60 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 70.6422 | 55.0459 | heterocephalus_glaber_female | ENSHGLP00000041441 | 75.4902 | 58.8235 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 75.2294 | 58.7156 | heterocephalus_glaber_female | ENSHGLP00000044063 | 82.8283 | 64.6465 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 76.1468 | 68.8073 | rhinopithecus_bieti | ENSRBIP00000017989 | 77.5701 | 70.0935 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 77.0642 | 69.7248 | rhinopithecus_bieti | ENSRBIP00000000155 | 89.3617 | 80.8511 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 85.3211 | 77.0642 | rhinopithecus_bieti | ENSRBIP00000023806 | 93 | 84 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 77.0642 | 67.8899 | rhinopithecus_bieti | ENSRBIP00000014578 | 84 | 74 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 84.4037 | 71.5596 | canis_lupus_dingo | ENSCAFP00020020001 | 82.8829 | 70.2703 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 56.8807 | 45.8716 | phascolarctos_cinereus | ENSPCIP00000036378 | 66.6667 | 53.7634 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 79.8165 | 64.2202 | nannospalax_galili | ENSNGAP00000015287 | 83.6538 | 67.3077 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 77.9817 | 64.2202 | bos_grunniens | ENSBGRP00000022656 | 72.6496 | 59.8291 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 86.2385 | 70.6422 | ictidomys_tridecemlineatus | ENSSTOP00000030751 | 84.6847 | 69.3694 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 79.8165 | 66.055 | cebus_imitator | ENSCCAP00000024652 | 70.1613 | 58.0645 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 87.156 | 70.6422 | urocitellus_parryii | ENSUPAP00010016123 | 85.5856 | 69.3694 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 78.8991 | 66.055 | moschus_moschiferus | ENSMMSP00000009229 | 72.8814 | 61.0169 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 80.7339 | 73.3945 | rhinopithecus_roxellana | ENSRROP00000035647 | 87.1287 | 79.2079 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 77.9817 | 69.7248 | rhinopithecus_roxellana | ENSRROP00000004212 | 88.5417 | 79.1667 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 77.0642 | 66.9725 | rhinopithecus_roxellana | ENSRROP00000023519 | 84 | 73 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 75.2294 | 65.1376 | propithecus_coquereli | ENSPCOP00000007800 | 70.6897 | 61.2069 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 49.5413 | 33.945 | petromyzon_marinus | ENSPMAP00000010284 | 60 | 41.1111 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 65.1376 | 45.8716 | callorhinchus_milii | ENSCMIP00000039507 | 70.297 | 49.505 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 62.3853 | 44.9541 | latimeria_chalumnae | ENSLACP00000015262 | 65.3846 | 47.1154 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 65.1376 | 46.789 | clupea_harengus | ENSCHAP00000037162 | 71 | 51 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 43.1193 | danio_rerio | ENSDARP00000127876 | 67.9612 | 45.6311 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 41.2844 | carassius_auratus | ENSCARP00000112211 | 68.6274 | 44.1176 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 42.2018 | carassius_auratus | ENSCARP00000125506 | 50.7353 | 33.8235 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 42.2018 | carassius_auratus | ENSCARP00000090778 | 37.5 | 25 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 60.5505 | 42.2018 | astyanax_mexicanus | ENSAMXP00000019246 | 66 | 46 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 62.3853 | 41.2844 | ictalurus_punctatus | ENSIPUP00000014984 | 49.635 | 32.8467 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 56.8807 | 39.4495 | esox_lucius | ENSELUP00000065350 | 60.7843 | 42.1569 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 55.9633 | 35.7798 | electrophorus_electricus | ENSEEEP00000047135 | 58.6538 | 37.5 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 58.7156 | 37.6147 | oncorhynchus_kisutch | ENSOKIP00005093928 | 42.3841 | 27.1523 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 61.4679 | 42.2018 | oncorhynchus_kisutch | ENSOKIP00005021084 | 60.9091 | 41.8182 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 61.4679 | 42.2018 | oncorhynchus_mykiss | ENSOMYP00000064261 | 67 | 46 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 57.7982 | 38.5321 | oncorhynchus_mykiss | ENSOMYP00000026408 | 38.8889 | 25.9259 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 42.2018 | salmo_salar | ENSSSAP00000006150 | 68.3168 | 45.5446 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 57.7982 | 35.7798 | salmo_salar | ENSSSAP00000139525 | 39.1304 | 24.2236 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 43.1193 | salmo_trutta | ENSSTUP00000052574 | 69.3069 | 46.5347 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 58.7156 | 39.4495 | salmo_trutta | ENSSTUP00000050324 | 39.2638 | 26.3804 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 55.0459 | 38.5321 | gadus_morhua | ENSGMOP00000018196 | 54.0541 | 37.8378 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 53.211 | 38.5321 | fundulus_heteroclitus | ENSFHEP00000003813 | 63.0435 | 45.6522 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 46.789 | poecilia_reticulata | ENSPREP00000017276 | 69 | 51 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 47.7064 | xiphophorus_maculatus | ENSXMAP00000015685 | 69 | 52 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 43.1193 | oryzias_latipes | ENSORLP00000013496 | 54.2636 | 36.4341 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 62.3853 | 42.2018 | cyclopterus_lumpus | ENSCLMP00005016557 | 68 | 46 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 43.1193 | oreochromis_niloticus | ENSONIP00000058245 | 70.7071 | 47.4747 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 61.4679 | 45.8716 | sparus_aurata | ENSSAUP00010025654 | 67 | 50 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 60.5505 | 43.1193 | lates_calcarifer | ENSLCAP00010000086 | 66 | 47 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 66.9725 | 46.789 | xenopus_tropicalis | ENSXETP00000060929 | 73.7374 | 51.5152 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 58.7156 | 44.9541 | chrysemys_picta_bellii | ENSCPBP00000038011 | 64.6465 | 49.4949 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 46.789 | sphenodon_punctatus | ENSSPUP00000012170 | 70.4082 | 52.0408 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 55.0459 | 37.6147 | notechis_scutatus | ENSNSUP00000000480 | 67.4157 | 46.0674 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 57.7982 | 40.367 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000000288 | 58.8785 | 41.1215 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 62.3853 | 36.6972 | gallus_gallus | ENSGALP00010006489 | 65.3846 | 38.4615 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 58.7156 | 35.7798 | serinus_canaria | ENSSCAP00000012361 | 60.3774 | 36.7925 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 60.5505 | 40.367 | parus_major | ENSPMJP00000022776 | 60 | 40 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 50.4587 | 36.6972 | anolis_carolinensis | ENSACAP00000037401 | 71.4286 | 51.9481 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 54.1284 | 37.6147 | naja_naja | ENSNNAP00000000250 | 55.1402 | 38.3178 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 51.3761 | 32.1101 | podarcis_muralis | ENSPMRP00000019077 | 54.3689 | 33.9806 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 58.7156 | 37.6147 | taeniopygia_guttata | ENSTGUP00000012702 | 60.3774 | 38.6792 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 43.1193 | erpetoichthys_calabaricus | ENSECRP00000030558 | 70.7071 | 47.4747 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 52.2936 | 29.3578 | kryptolebias_marmoratus | ENSKMAP00000002667 | 52.7778 | 29.6296 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 48.6239 | 39.4495 | salvator_merianae | ENSSMRP00000006324 | 42.7419 | 34.6774 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 58.7156 | 44.0367 | salvator_merianae | ENSSMRP00000016000 | 65.9794 | 49.4845 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 66.055 | 45.8716 | oryzias_javanicus | ENSOJAP00000043689 | 72 | 50 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 42.2018 | amphilophus_citrinellus | ENSACIP00000029822 | 69.3069 | 45.5446 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 61.4679 | 42.2018 | dicentrarchus_labrax | ENSDLAP00005007975 | 67.6768 | 46.4646 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 59.633 | 39.4495 | amphiprion_percula | ENSAPEP00000022323 | 59.0909 | 39.0909 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 59.633 | 39.4495 | amphiprion_percula | ENSAPEP00000022057 | 59.0909 | 39.0909 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 43.1193 | oryzias_sinensis | ENSOSIP00000023178 | 70 | 47 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 69.7248 | 52.2936 | gopherus_evgoodei | ENSGEVP00005000211 | 72.381 | 54.2857 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 66.9725 | 47.7064 | chelonoidis_abingdonii | ENSCABP00000031234 | 69.5238 | 49.5238 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 44.9541 | seriola_dumerili | ENSSDUP00000030374 | 45.3947 | 32.2368 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 42.2018 | cottoperca_gobio | ENSCGOP00000027317 | 70 | 46 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 56.8807 | 38.5321 | gasterosteus_aculeatus | ENSGACP00000026602 | 72.093 | 48.8372 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 55.0459 | 41.2844 | cynoglossus_semilaevis | ENSCSEP00000024676 | 61.8557 | 46.3918 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 46.789 | poecilia_formosa | ENSPFOP00000006669 | 69 | 51 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 41.2844 | myripristis_murdjan | ENSMMDP00005012218 | 66.0377 | 42.4528 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 65.1376 | 44.9541 | sander_lucioperca | ENSSLUP00000039664 | 50.3546 | 34.7518 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 59.633 | 33.0275 | strigops_habroptila | ENSSHBP00005004821 | 61.3208 | 33.9623 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 41.2844 | amphiprion_ocellaris | ENSAOCP00000021505 | 70.7071 | 45.4545 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 59.633 | 45.8716 | terrapene_carolina_triunguis | ENSTMTP00000023696 | 74.7126 | 57.4713 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 44.0367 | hucho_hucho | ENSHHUP00000076223 | 69.3069 | 47.5248 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 56.8807 | 37.6147 | betta_splendens | ENSBSLP00000049700 | 59.6154 | 39.4231 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 66.9725 | 46.789 | pygocentrus_nattereri | ENSPNAP00000002197 | 73.7374 | 51.5152 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 42.2018 | anabas_testudineus | ENSATEP00000019406 | 70 | 46 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 44.9541 | seriola_lalandi_dorsalis | ENSSLDP00000030006 | 62.7273 | 44.5455 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 60.5505 | 41.2844 | labrus_bergylta | ENSLBEP00000009776 | 66 | 45 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 62.3853 | 44.0367 | mastacembelus_armatus | ENSMAMP00000023903 | 31.1927 | 22.0183 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 42.2018 | stegastes_partitus | ENSSPAP00000017824 | 70 | 46 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 57.7982 | 38.5321 | oncorhynchus_tshawytscha | ENSOTSP00005023293 | 38.1818 | 25.4545 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 61.4679 | 41.2844 | oncorhynchus_tshawytscha | ENSOTSP00005053266 | 67 | 45 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 65.1376 | 41.2844 | acanthochromis_polyacanthus | ENSAPOP00000000135 | 71 | 45 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 57.7982 | 35.7798 | aquila_chrysaetos_chrysaetos | ENSACCP00020001456 | 24.6094 | 15.2344 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 50.4587 | 37.6147 | nothobranchius_furzeri | ENSNFUP00015031128 | 49.1071 | 36.6071 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 61.4679 | 44.9541 | cyprinodon_variegatus | ENSCVAP00000010379 | 66.3366 | 48.5149 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 66.055 | 44.9541 | denticeps_clupeoides | ENSDCDP00000007517 | 72 | 49 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 65.1376 | 46.789 | larimichthys_crocea | ENSLCRP00005001141 | 71 | 51 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 62.3853 | 43.1193 | takifugu_rubripes | ENSTRUP00000050378 | 48.227 | 33.3333 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 44.0367 | hippocampus_comes | ENSHCOP00000005167 | 70.7071 | 48.4848 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 42.2018 | cyprinus_carpio_carpio | ENSCCRP00000087107 | 67.6471 | 45.098 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 64.2202 | 43.1193 | cyprinus_carpio_carpio | ENSCCRP00000120599 | 67.3077 | 45.1923 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 61.4679 | 41.2844 | coturnix_japonica | ENSCJPP00005024494 | 59.292 | 39.823 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 46.789 | poecilia_latipinna | ENSPLAP00000008557 | 69 | 51 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 41.2844 | sinocyclocheilus_grahami | ENSSGRP00000102346 | 67.6471 | 44.1176 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 43.1193 | sinocyclocheilus_grahami | ENSSGRP00000073068 | 65.0943 | 44.3396 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 63.3028 | 42.2018 | maylandia_zebra | ENSMZEP00005005955 | 69.697 | 46.4646 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 55.9633 | 38.5321 | paramormyrops_kingsleyae | ENSPKIP00000026282 | 60.396 | 41.5842 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 61.4679 | 44.0367 | scleropages_formosus | ENSSFOP00015068909 | 67.6768 | 48.4848 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 59.633 | 44.9541 | leptobrachium_leishanense | ENSLLEP00000047032 | 69.8925 | 52.6882 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 55.0459 | 34.8624 | scophthalmus_maximus | ENSSMAP00000021090 | 57.6923 | 36.5385 |
| ENSG00000119705 | homo_sapiens | ENSP00000450909 | 65.1376 | 44.9541 | oryzias_melastigma | ENSOMEP00000033363 | 71 | 49 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000961 | involved_in negative regulation of mitochondrial RNA catabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001669 | located_in acrosomal vesicle | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005739 | located_in mitochondrion | 2 | HDA,IDA | Homo_sapiens(9606) | Component |
| GO:0007286 | involved_in spermatid development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007338 | involved_in single fertilization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0030317 | involved_in flagellated sperm motility | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0036126 | located_in sperm flagellum | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0048471 | located_in perinuclear region of cytoplasm | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0070584 | involved_in mitochondrion morphogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1990904 | part_of ribonucleoprotein complex | 1 | IEA | Homo_sapiens(9606) | Component |