RBP Type
Non-canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
N.A.
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000117602 Gene ID
11123 Accession
3042
Symbol
RCAN3
Alias
RCN3;MCIP3;hRCN3;DSCR1L2
Full Name
RCAN family member 3
Status
Confidence
Length
38690 bases
Strand
Plus strand
Position
1 : 24502351 - 24541040
RNA binding domain
N.A.
Summary
Enables phosphatase binding activity and troponin I binding activity. Predicted to be involved in calcium-mediated signaling. Predicted to be active in cytoplasm and nucleus. [provided by Alliance of Genome Resources, Apr 2022]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
34529546 A systematic review and meta-analysis of imaging genetics studies of specific reading disorder.
Tina Thomas
2021-05-01
Cognitive neuropsychology
Name
Transcript ID
bp
Protein
Translation ID
RCAN3-207
ENST00000538532
2421
241aa
ENSP00000445401
RCAN3-209
ENST00000618490
2386
116aa
ENSP00000484519
RCAN3-202
ENST00000374395
6863
241aa
ENSP00000363516
RCAN3-205
ENST00000436717
2574
183aa
ENSP00000414447
RCAN3-208
ENST00000616511
2527
116aa
ENSP00000478174
RCAN3-204
ENST00000425530
997
231aa
ENSP00000409540
RCAN3-203
ENST00000412742
667
173aa
ENSP00000391912
RCAN3-210
ENST00000630217
658
183aa
ENSP00000486836
RCAN3-201
ENST00000374393
382
115aa
ENSP00000363514
RCAN3-206
ENST00000482807
497
108aa
ENSP00000437338
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
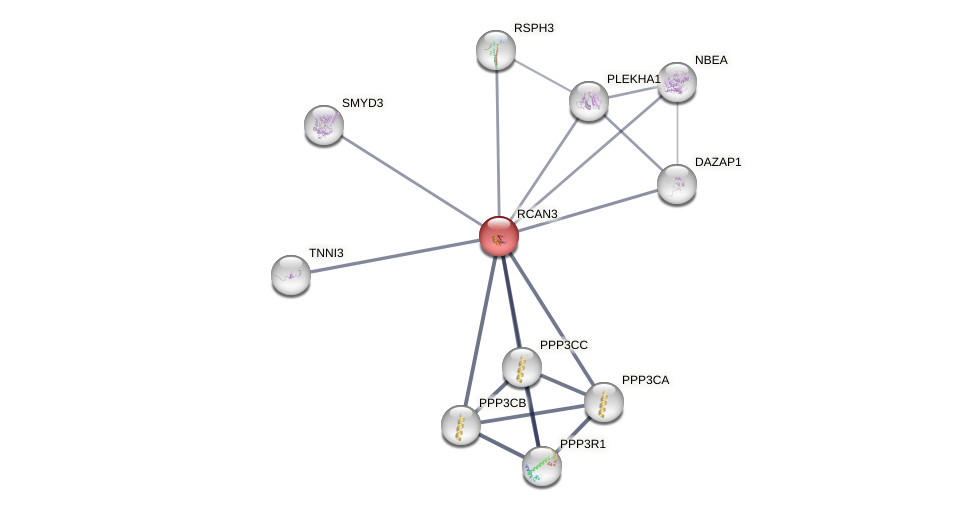
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0003723 enables RNA binding 1 TAS Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 is_active_in nucleus 1 IBA Homo_sapiens(9606) Component GO:0005737 is_active_in cytoplasm 1 IBA Homo_sapiens(9606) Component GO:0005829 located_in cytosol 1 TAS Homo_sapiens(9606) Component GO:0008597 enables calcium-dependent protein serine/threonine phosphatase regulator activity 1 IBA Homo_sapiens(9606) Function GO:0009653 involved_in anatomical structure morphogenesis 1 TAS Homo_sapiens(9606) Process GO:0019722 involved_in calcium-mediated signaling 1 IBA Homo_sapiens(9606) Process GO:0019902 enables phosphatase binding 1 IPI Homo_sapiens(9606) Function GO:0031013 enables troponin I binding 1 IPI Homo_sapiens(9606) Function
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.