RBP Type
Canonical_RBPs
Diseases
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
TF
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000117595 Gene ID
3664 Accession
6121
Symbol
IRF6
Alias
LPS;PIT;PPS;VWS;OFC6;PPS1;VWS1
Full Name
interferon regulatory factor 6
Status
Confidence
Length
20559 bases
Strand
Minus strand
Position
1 : 209785617 - 209806175
RNA binding domain
IRF-3
Summary
This gene encodes a member of the interferon regulatory transcription factor (IRF) family. Family members share a highly-conserved N-terminal helix-turn-helix DNA-binding domain and a less conserved C-terminal protein-binding domain. The encoded protein may be a transcriptional activator. Mutations in this gene can cause van der Woude syndrome and popliteal pterygium syndrome. Mutations in this gene are also associated with non-syndromic orofacial cleft type 6. Alternate splicing results in multiple transcript variants.[provided by RefSeq, May 2011]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000440532
IRF-3
PF10401 1.4e-68
1
ENSP00000355988
IRF-3
PF10401 2.3e-68
1
ENSP00000403855
IRF-3
PF10401 2.7e-09
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
32527996 Lin28A promotes IRF6-regulated aerobic glycolysis in glioma cells by stabilizing SNHG14.
Jinjing Lu
2020-06-11
Cell death & disease
Name
Transcript ID
bp
Protein
Translation ID
IRF6-201
ENST00000367021
4478
467aa
ENSP00000355988
IRF6-204
ENST00000542854
4256
372aa
ENSP00000440532
IRF6-206
ENST00000696134
4558
235aa
ENSP00000512427
IRF6-205
ENST00000643798
1697
179aa
ENSP00000496669
IRF6-202
ENST00000456314
1027
276aa
ENSP00000403855
IRF6-203
ENST00000464698
596
No protein
-
Pathway ID
Pathway Name
Source
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
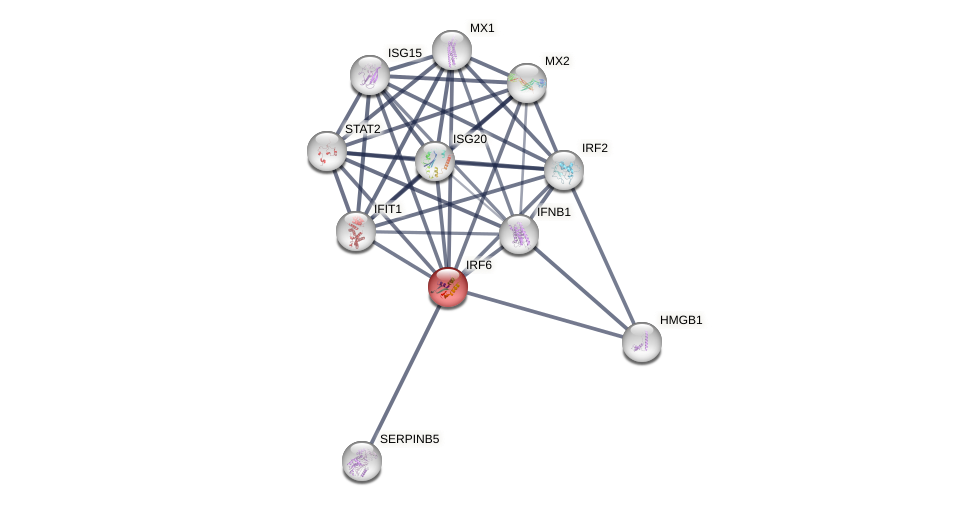
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000785 part_of chromatin 1 ISA Homo_sapiens(9606) Component GO:0000978 enables RNA polymerase II cis-regulatory region sequence-specific DNA binding 1 IBA Homo_sapiens(9606) Function GO:0000981 enables DNA-binding transcription factor activity, RNA polymerase II-specific 2 IBA,ISA Homo_sapiens(9606) Function GO:0001228 enables DNA-binding transcription activator activity, RNA polymerase II-specific 1 IEA Homo_sapiens(9606) Function GO:0002376 involved_in immune system process 1 IBA Homo_sapiens(9606) Process GO:0003677 enables DNA binding 1 ISS Homo_sapiens(9606) Function GO:0003700 enables DNA-binding transcription factor activity 1 ISS Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 is_active_in nucleus 1 IBA Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 1 IDA Homo_sapiens(9606) Component GO:0005737 located_in cytoplasm 1 IDA Homo_sapiens(9606) Component GO:0005829 located_in cytosol 2 IDA,TAS Homo_sapiens(9606) Component GO:0006357 involved_in regulation of transcription by RNA polymerase II 1 IBA Homo_sapiens(9606) Process GO:0008285 involved_in negative regulation of cell population proliferation 1 IDA Homo_sapiens(9606) Process GO:0010839 involved_in negative regulation of keratinocyte proliferation 1 IEA Homo_sapiens(9606) Process GO:0030054 located_in cell junction 1 IDA Homo_sapiens(9606) Component GO:0030216 involved_in keratinocyte differentiation 1 IEA Homo_sapiens(9606) Process GO:0043565 enables sequence-specific DNA binding 1 IDA Homo_sapiens(9606) Function GO:0043616 involved_in keratinocyte proliferation 1 IEA Homo_sapiens(9606) Process GO:0045893 involved_in positive regulation of DNA-templated transcription 1 ISS Homo_sapiens(9606) Process GO:0045944 involved_in positive regulation of transcription by RNA polymerase II 1 IDA Homo_sapiens(9606) Process GO:0048468 involved_in cell development 1 IEA Homo_sapiens(9606) Process GO:0060021 involved_in roof of mouth development 1 IEA Homo_sapiens(9606) Process GO:0060173 involved_in limb development 1 IEA Homo_sapiens(9606) Process GO:0060644 involved_in mammary gland epithelial cell differentiation 1 ISS Homo_sapiens(9606) Process GO:0070062 located_in extracellular exosome 1 HDA Homo_sapiens(9606) Component GO:0072089 involved_in stem cell proliferation 1 IEA Homo_sapiens(9606) Process GO:1904888 involved_in cranial skeletal system development 1 IEA Homo_sapiens(9606) Process GO:1990837 enables sequence-specific double-stranded DNA binding 1 IDA Homo_sapiens(9606) Function GO:2000647 involved_in negative regulation of stem cell proliferation 1 IEA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.