RBP Type
Non-canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
E3 ligase
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000100023 Gene ID
23759 Accession
9261
Symbol
PPIL2
Alias
CYC4;CYP60;UBOX7;Cyp-60;hCyP-60
Full Name
peptidylprolyl isomerase like 2
Status
Confidence
Length
34016 bases
Strand
Plus strand
Position
22 : 21666000 - 21700015
RNA binding domain
N.A.
Summary
This gene is a member of the cyclophilin family of peptidylprolyl isomerases. The cyclophilins are a highly conserved ubiquitous family, members of which play an important role in protein folding, immunosuppression by cyclosporin A, and infection of HIV-1 virions. This protein interacts with the proteinase inhibitor eglin c and is localized in the nucleus. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Dec 2015]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
PPIL2-233
ENST00000680463
1996
No protein
-
PPIL2-230
ENST00000680393
2840
520aa
ENSP00000506542
PPIL2-214
ENST00000679479
2856
No protein
-
PPIL2-215
ENST00000679534
3507
520aa
ENSP00000504893
PPIL2-222
ENST00000679827
2692
510aa
ENSP00000504929
PPIL2-246
ENST00000681956
4168
520aa
ENSP00000506158
PPIL2-240
ENST00000681338
3025
573aa
ENSP00000505107
PPIL2-225
ENST00000680061
3080
520aa
ENSP00000505910
PPIL2-218
ENST00000679580
3238
415aa
ENSP00000506723
PPIL2-239
ENST00000681286
3214
No protein
-
PPIL2-216
ENST00000679540
3005
520aa
ENSP00000506328
PPIL2-221
ENST00000679795
4051
520aa
ENSP00000506144
PPIL2-231
ENST00000680426
3138
No protein
-
PPIL2-210
ENST00000496819
2411
No protein
-
PPIL2-211
ENST00000498109
3277
No protein
-
PPIL2-201
ENST00000335025
4929
520aa
ENSP00000334553
PPIL2-242
ENST00000681394
4115
305aa
ENSP00000506123
PPIL2-202
ENST00000398831
4068
520aa
ENSP00000381812
PPIL2-213
ENST00000679477
3237
549aa
ENSP00000505978
PPIL2-212
ENST00000626352
2625
527aa
ENSP00000486725
PPIL2-206
ENST00000458567
557
160aa
ENSP00000395181
PPIL2-204
ENST00000417788
4213
41aa
ENSP00000406745
PPIL2-219
ENST00000679586
1812
No protein
-
PPIL2-223
ENST00000679857
2503
No protein
-
PPIL2-236
ENST00000680962
3854
305aa
ENSP00000505778
PPIL2-245
ENST00000681791
3070
250aa
ENSP00000505019
PPIL2-220
ENST00000679692
3235
305aa
ENSP00000504937
PPIL2-226
ENST00000680094
3343
614aa
ENSP00000506368
PPIL2-208
ENST00000484439
863
No protein
-
PPIL2-232
ENST00000680434
3381
43aa
ENSP00000505533
PPIL2-227
ENST00000680109
3076
499aa
ENSP00000506219
PPIL2-203
ENST00000406385
3875
520aa
ENSP00000384299
PPIL2-224
ENST00000680022
1523
No protein
-
PPIL2-238
ENST00000681137
3156
101aa
ENSP00000505216
PPIL2-235
ENST00000680860
3062
342aa
ENSP00000506196
PPIL2-229
ENST00000680183
3840
No protein
-
PPIL2-209
ENST00000485930
149
No protein
-
PPIL2-241
ENST00000681353
587
No protein
-
PPIL2-205
ENST00000446951
1926
No protein
-
PPIL2-244
ENST00000681735
3391
No protein
-
PPIL2-217
ENST00000679564
2509
No protein
-
PPIL2-228
ENST00000680113
3672
No protein
-
PPIL2-234
ENST00000680573
2972
No protein
-
PPIL2-243
ENST00000681612
562
No protein
-
PPIL2-207
ENST00000462188
1735
No protein
-
PPIL2-237
ENST00000681027
3437
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
21317 Choice Behavior 4.136e-005 - 21326 Choice Behavior 4.136e-005 - 40234 Platelet Function Tests 5.7000000E-007 - 70553 Electrocardiography 2E-6 24324551 70554 Trypanosoma cruzi 2E-6 24324551
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
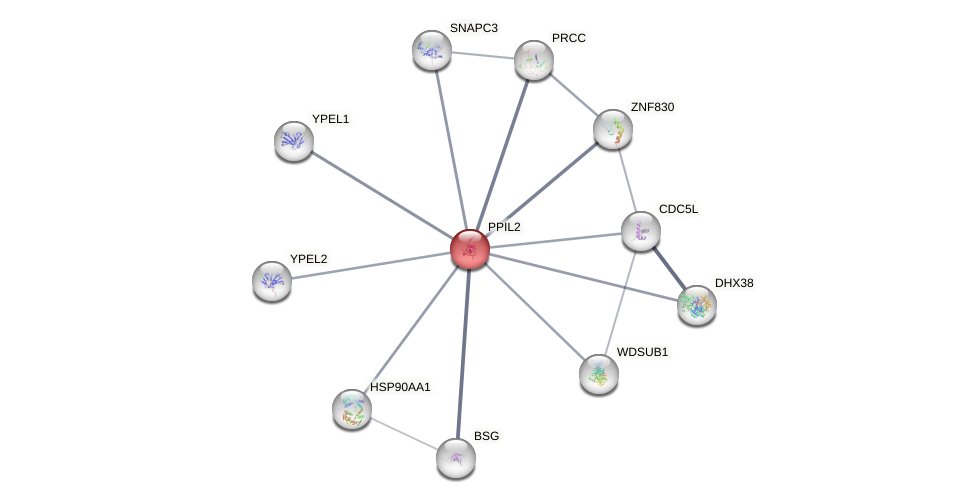
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000209 involved_in protein polyubiquitination 2 IBA,IDA Homo_sapiens(9606) Process GO:0000413 involved_in protein peptidyl-prolyl isomerization 1 IEA Homo_sapiens(9606) Process GO:0003755 NOT enables peptidyl-prolyl cis-trans isomerase activity 1 IDA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 located_in nucleus 2 IDA,TAS Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 2 IDA,TAS Homo_sapiens(9606) Component GO:0005737 located_in cytoplasm 1 IDA Homo_sapiens(9606) Component GO:0005796 located_in Golgi lumen 1 TAS Homo_sapiens(9606) Component GO:0005886 located_in plasma membrane 1 IDA Homo_sapiens(9606) Component GO:0006397 involved_in mRNA processing 1 IEA Homo_sapiens(9606) Process GO:0006457 involved_in protein folding 1 IEA Homo_sapiens(9606) Process GO:0008380 involved_in RNA splicing 1 IEA Homo_sapiens(9606) Process GO:0016018 NOT enables cyclosporin A binding 1 IDA Homo_sapiens(9606) Function GO:0034450 enables ubiquitin-ubiquitin ligase activity 1 IDA Homo_sapiens(9606) Function GO:0061630 enables ubiquitin protein ligase activity 2 IBA,IDA Homo_sapiens(9606) Function GO:0071013 part_of catalytic step 2 spliceosome 1 IBA Homo_sapiens(9606) Component GO:0072659 involved_in protein localization to plasma membrane 1 IMP Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.