RBP Type
Non-canonical_RBPs
Diseases
Flaviviruses SARS-COV-2 Dengue Zika
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
N.A.
Localizations
Stress granucle
BulkPerturb-seq
Ensembl ID ENSG00000087302 Gene ID
51637 Accession
23169
Symbol
RTRAF
Alias
CLE;CLE7;hCLE;CGI99;RLLM1;hCLE1;CGI-99;LCRP369;C14orf166
Full Name
RNA transcription, translation and transport factor
Status
Confidence
Length
21181 bases
Strand
Plus strand
Position
14 : 51989514 - 52010694
RNA binding domain
N.A.
Summary
Enables RNA binding activity; RNA polymerase II complex binding activity; and identical protein binding activity. Involved in negative regulation of protein kinase activity; positive regulation of transcription by RNA polymerase II; and tRNA splicing, via endonucleolytic cleavage and ligation. Located in microtubule cytoskeleton; nucleoplasm; and perinuclear region of cytoplasm. Part of tRNA-splicing ligase complex. [provided by Alliance of Genome Resources, Apr 2022]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
RTRAF-201
ENST00000261700
7007
244aa
ENSP00000261700
RTRAF-207
ENST00000557553
557
68aa
ENSP00000451751
RTRAF-206
ENST00000556760
851
193aa
ENSP00000451715
RTRAF-202
ENST00000553362
683
129aa
ENSP00000451153
RTRAF-204
ENST00000553707
822
No protein
-
RTRAF-205
ENST00000555319
578
No protein
-
RTRAF-203
ENST00000553479
562
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
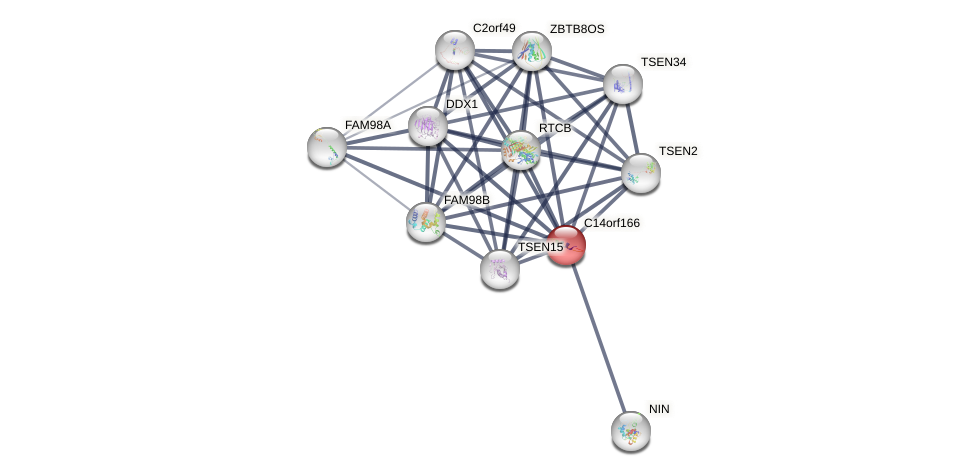
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000993 enables RNA polymerase II complex binding 1 IDA Homo_sapiens(9606) Function GO:0003723 enables RNA binding 3 HDA,IBA,IDA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 located_in nucleus 2 IDA,NAS Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 2 IDA,TAS Homo_sapiens(9606) Component GO:0005737 located_in cytoplasm 2 IDA,NAS Homo_sapiens(9606) Component GO:0005813 located_in centrosome 1 IDA Homo_sapiens(9606) Component GO:0005829 located_in cytosol 1 IEA Homo_sapiens(9606) Component GO:0006388 involved_in tRNA splicing, via endonucleolytic cleavage and ligation 3 IBA,IMP,NAS Homo_sapiens(9606) Process GO:0006469 involved_in negative regulation of protein kinase activity 1 IMP Homo_sapiens(9606) Process GO:0042802 enables identical protein binding 1 IPI Homo_sapiens(9606) Function GO:0045944 involved_in positive regulation of transcription by RNA polymerase II 1 IMP Homo_sapiens(9606) Process GO:0048471 located_in perinuclear region of cytoplasm 1 IDA Homo_sapiens(9606) Component GO:0050658 involved_in RNA transport 1 TAS Homo_sapiens(9606) Process GO:0072669 part_of tRNA-splicing ligase complex 3 IBA,IDA,NAS Homo_sapiens(9606) Component GO:0072686 located_in mitotic spindle 1 IDA Homo_sapiens(9606) Component
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.