RBP Type
Non-canonical_RBPs
Diseases
N.A.
Drug
Main interacting RNAs
N.A.
Moonlighting functions
Kinase
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000071242 Gene ID
6196 Accession
10431
Symbol
RPS6KA2
Alias
RSK;HU-2;RSK3;p90RSK2;p90-RSK3;pp90RSK3;MAPKAPK1C;S6K-alpha;S6K-alpha2
Full Name
ribosomal protein S6 kinase A2
Status
Confidence
Length
497088 bases
Strand
Minus strand
Position
6 : 166409364 - 166906451
RNA binding domain
N.A.
Summary
This gene encodes a member of the RSK (ribosomal S6 kinase) family of serine/threonine kinases. This kinase contains two non-identical kinase catalytic domains and phosphorylates various substrates, including members of the mitogen-activated kinase (MAPK) signalling pathway. The activity of this protein has been implicated in controlling cell growth and differentiation. Alternative splice variants, encoding different isoforms, have been characterized. [provided by RefSeq, Jan 2016]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
447 Maximal Midexpiratory Flow Rate 2.30381047463241E-6 17903307 483 Respiratory Function Tests 3.27031057202709E-10 17903307 573 Maximal Midexpiratory Flow Rate 4.71311162658954E-6 17903307 616 Respiratory Function Tests 6.39834499148856E-9 17903307 15615 Heart Failure 5.3537282E-004 - 21955 Personality 2.66e-005 - 28020 Diabetes Mellitus 2.0960000E-005 - 28031 Diabetes Mellitus 2.2780000E-005 - 31641 Platelet Count 2.5980000E-005 - 32053 Fetal Hemoglobin 1.6100000E-005 20018918 32084 Fetal Hemoglobin 7.2100000E-006 20018918 36186 Platelet Function Tests 1.6100000E-006 - 36212 Platelet Function Tests 6.8900000E-008 - 36216 Platelet Function Tests 3.6200000E-007 - 45100 Platelet Function Tests 2.4505074E-006 - 51132 Child Development Disorders, Pervasive 4.6259000E-005 - 52389 Child Development Disorders, Pervasive 2.4000000E-005 - 52600 Blood Urea Nitrogen 5.5580000E-005 - 54676 Creatine Kinase 5.0640000E-005 - 94783 Blood Pressure 8E-7 25189868 94784 Smoking 8E-7 25189868 100749 Dental Caries 1E-7 23259602
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
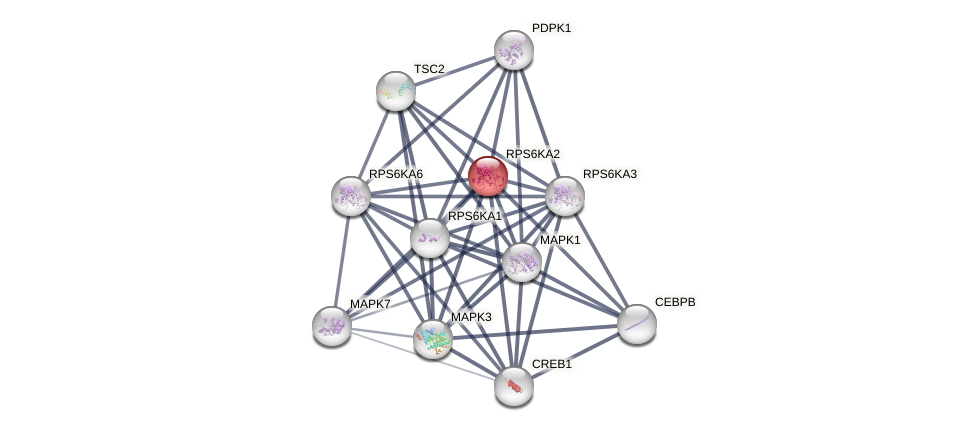
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000287 enables magnesium ion binding 1 IEA Homo_sapiens(9606) Function GO:0001556 involved_in oocyte maturation 1 IEA Homo_sapiens(9606) Process GO:0002035 involved_in brain renin-angiotensin system 1 IEA Homo_sapiens(9606) Process GO:0004674 enables protein serine/threonine kinase activity 1 TAS Homo_sapiens(9606) Function GO:0004711 enables ribosomal protein S6 kinase activity 1 IBA Homo_sapiens(9606) Function GO:0004712 enables protein serine/threonine/tyrosine kinase activity 1 IDA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005524 enables ATP binding 1 IEA Homo_sapiens(9606) Function GO:0005634 located_in nucleus 1 IDA Homo_sapiens(9606) Component GO:0005654 is_active_in nucleoplasm 3 IBA,IDA,TAS Homo_sapiens(9606) Component GO:0005737 is_active_in cytoplasm 2 IBA,IDA Homo_sapiens(9606) Component GO:0005829 located_in cytosol 1 TAS Homo_sapiens(9606) Component GO:0007165 involved_in signal transduction 1 NAS Homo_sapiens(9606) Process GO:0007268 involved_in chemical synaptic transmission 1 TAS Homo_sapiens(9606) Process GO:0007507 involved_in heart development 1 IEA Homo_sapiens(9606) Process GO:0008285 involved_in negative regulation of cell population proliferation 1 IDA Homo_sapiens(9606) Process GO:0010628 involved_in positive regulation of gene expression 1 IEA Homo_sapiens(9606) Process GO:0010659 involved_in cardiac muscle cell apoptotic process 1 IEA Homo_sapiens(9606) Process GO:0018105 involved_in peptidyl-serine phosphorylation 1 IBA Homo_sapiens(9606) Process GO:0035556 involved_in intracellular signal transduction 1 IEA Homo_sapiens(9606) Process GO:0043065 involved_in positive regulation of apoptotic process 1 IDA Homo_sapiens(9606) Process GO:0045202 located_in synapse 1 IEA Homo_sapiens(9606) Component GO:0045786 involved_in negative regulation of cell cycle 1 IDA Homo_sapiens(9606) Process GO:0045835 involved_in negative regulation of meiotic nuclear division 1 IEA Homo_sapiens(9606) Process GO:0060047 involved_in heart contraction 1 IEA Homo_sapiens(9606) Process GO:0070613 involved_in regulation of protein processing 1 IEA Homo_sapiens(9606) Process GO:0071322 involved_in cellular response to carbohydrate stimulus 1 IEA Homo_sapiens(9606) Process GO:0072687 located_in meiotic spindle 1 IEA Homo_sapiens(9606) Component GO:0106310 enables protein serine kinase activity 1 IEA Homo_sapiens(9606) Function
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.