Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | CancerCHIKVSARS-COV-OC43FlavivirusesHIVRVSARS-COV-2SINVDengueZika |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | TF |
| Localizations | Stress granucleP-body |
| BulkPerturb-seq | DataSet_02_121 DataSet_02_122 |
Description
| Ensembl ID | ENSG00000065978 | Gene ID | 4904 | Accession | 8014 |
| Symbol | YBX1 | Alias | YB1;BP-8;CSDB;DBPB;YB-1;CBF-A;CSDA2;EFI-A;NSEP1;NSEP-1;MDR-NF1 | Full Name | Y-box binding protein 1 |
| Status | Confidence | Length | 21388 bases | Strand | Plus strand |
| Position | 1 : 42682418 - 42703805 | RNA binding domain | CSD | ||
| Summary | This gene encodes a highly conserved cold shock domain protein that has broad nucleic acid binding properties. The encoded protein functions as both a DNA and RNA binding protein and has been implicated in numerous cellular processes including regulation of transcription and translation, pre-mRNA splicing, DNA reparation and mRNA packaging. This protein is also a component of messenger ribonucleoprotein (mRNP) complexes and may have a role in microRNA processing. This protein can be secreted through non-classical pathways and functions as an extracellular mitogen. Aberrant expression of the gene is associated with cancer proliferation in numerous tissues. This gene may be a prognostic marker for poor outcome and drug resistance in certain cancers. Alternate splicing results in multiple transcript variants. Pseudogenes of this gene are found on multiple chromosomes. [provided by RefSeq, Sep 2015] | ||||
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 37378715 | Translational control of Ybx1 expression regulates cardiac function in response to pressure overload in vivo. | Eshita Varma | 2023-06-28 | Basic research in cardiology |
| 36969033 | The lncRNA epigenetics: The significance of m6A and m5C lncRNA modifications in cancer. | Ylenia Vincenza Cusenza | 2023-01-01 | Frontiers in oncology |
| 31358880 | Altered transcriptional regulatory proteins in glioblastoma and YBX1 as a potential regulator of tumor invasion. | Kumar Manoj Gupta | 2019-07-29 | Scientific reports |
| 26843810 | Beyond the Ribosome: Extra-translational Functions of tRNA Fragments. | W Kevin Diebel | 2016-01-01 | Biomarker insights |
| 29808089 | Noncoding RNAs Carried by Extracellular Vesicles in Endocrine Diseases. | C A Margherita Pomatto | 2018-01-01 | International journal of endocrinology |
| 27559612 | Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. | J Matthew Shurtleff | 2016-08-25 | eLife |
| 29712925 | The RNA-binding protein YBX1 regulates epidermal progenitors at a posttranscriptional level. | Eunjeong Kwon | 2018-04-30 | Nature communications |
| 28257101 | Exosomes: From Garbage Bins to Promising Therapeutic Targets. | Mohammed M H | 2017-03-02 | International journal of molecular sciences |
| 31355251 | Whole Genome Analysis and Prognostic Model Construction Based on Alternative Splicing Events in Endometrial Cancer. | Caixia Wang | 2019-01-01 | BioMed research international |
| 25784054 | The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. | Guramrit Singh | 2015-01-01 | Annual review of biochemistry |
| 35961977 | A miR-34a-guided, tRNAiMet-derived, piR_019752-like fragment (tRiMetF31) suppresses migration and angiogenesis of breast cancer cells via targeting PFKFB3. | Bo Wang | 2022-08-12 | Cell death discovery |
| 36387119 | Plasma exosomal tRNA-derived fragments as diagnostic biomarkers in non-small cell lung cancer. | Baibing Zheng | 2022-01-01 | Frontiers in oncology |
| 29685978 | Interplay between long non-coding RNAs and epigenetic machinery: emerging targets in cancer? | J David Hanly | 2018-06-05 | Philosophical transactions of the Royal Society of London. Series B, Biological sciences |
| 32585856 | YBX1 Indirectly Targets Heterochromatin-Repressed Inflammatory Response-Related Apoptosis Genes through Regulating CBX5 mRNA. | Andreas Kloetgen | 2020-06-23 | International journal of molecular sciences |
| 34533860 | LncRNA BASP1-AS1 interacts with YBX1 to regulate Notch transcription and drives the malignancy of melanoma. | YaLing Li | 2021-11-01 | Cancer science |
| 33446492 | Comprehensive annotation and characterization of planarian tRNA and tRNA-derived fragments (tRFs). | Vairavan Lakshmanan | 2021-04-01 | RNA (New York, N.Y.) |
| 32947897 | Long Non-Coding RNAs as Strategic Molecules to Augment the Radiation Therapy in Esophageal Squamous Cell Carcinoma. | Uttam Sharma | 2020-09-16 | International journal of molecular sciences |
| 34465866 | YBX1 mediates translation of oncogenic transcripts to control cell competition in AML. | Florian Perner | 2022-02-01 | Leukemia |
| 28872613 | Roles of Long Noncoding RNAs in Recurrence and Metastasis of Radiotherapy-Resistant Cancer Stem Cells. | Hsiang-Cheng Chi | 2017-09-05 | International journal of molecular sciences |
| 37502906 | Analysis of small EV proteomes reveals unique functional protein networks regulated by VAP-A. | Bahnisikha Barman | 2023-07-19 | bioRxiv : the preprint server for biology |
| 36389669 | RNA m5C regulator-mediated modification patterns and the cross-talk between tumor microenvironment infiltration in gastric cancer. | Qiang Zhang | 2022-01-01 | Frontiers in immunology |
| 32178290 | Aurora Kinase A-YBX1 Synergy Fuels Aggressive Oncogenic Phenotypes and Chemoresistance in Castration-Resistant Prostate Cancer. | Kumar Nikhil | 2020-03-12 | Cancers |
| 36642732 | Transcriptional and post-transcriptional control of autophagy and adipogenesis by YBX1. | Ruifan Wu | 2023-01-16 | Cell death & disease |
| 25959498 | lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. | Yongchao Liu | 2015-05-11 | Scientific reports |
| 36533073 | Insights into the role of long non-coding RNAs in DNA methylation mediated transcriptional regulation. | Zhen Yang | 2022-01-01 | Frontiers in molecular biosciences |
| 29388131 | Extracellular RNAs: A New Awareness of Old Perspectives. | Noah Sadik | 2018-01-01 | Methods in molecular biology (Clifton, N.J.) |
| 29180470 | ERα Binding by Transcription Factors NFIB and YBX1 Enables FGFR2 Signaling to Modulate Estrogen Responsiveness in Breast Cancer. | M Thomas Campbell | 2018-01-15 | Cancer research |
| 20428086 | Pharmacogenomic identification of c-Myc/Max-regulated genes associated with cytotoxicity of artesunate towards human colon, ovarian and lung cancer cell lines. | Serkan Sertel | 2010-04-22 | Molecules (Basel, Switzerland) |
| 32354721 | 5-methylcytosine modification of an Epstein-Barr virus noncoding RNA decreases its stability. | A Belle Henry | 2020-08-01 | RNA (New York, N.Y.) |
| 27015120 | A comprehensive repertoire of tRNA-derived fragments in prostate cancer. | Michael Olvedy | 2016-04-26 | Oncotarget |
| 26307974 | Regulatory Roles of Non-Coding RNAs in Colorectal Cancer. | Jun Wang | 2015-08-21 | International journal of molecular sciences |
| 33391635 | RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? | Fabrizio Fabbiano | 2020-12-01 | Journal of extracellular vesicles |
| 36881772 | LncRNA HOTAIRM1 functions in DNA double-strand break repair via its association with DNA repair and mRNA surveillance factors. | Tzu-Wei Chuang | 2023-04-24 | Nucleic acids research |
| 34660788 | Methylation Modification, Alternative Splicing, and Noncoding RNA Play a Role in Cancer Metastasis through Epigenetic Regulation. | Bin Yu | 2021-01-01 | BioMed research international |
| 36754932 | YBX1/lncRNA SBF2-AS1 interaction regulates proliferation and tamoxifen sensitivity via PI3K/AKT/MTOR signaling in breast cancer cells. | A Shaharbhanu Hussain | 2023-04-01 | Molecular biology reports |
| 35109938 | YBX1 mediates alternative splicing and maternal mRNA decay during pre-implantation development. | Mingtian Deng | 2022-02-02 | Cell & bioscience |
| 35861369 | Genetic variants in RNA m5 C modification genes associated with survival and chemotherapy efficacy of colorectal cancer. | Silu Chen | 2023-01-01 | Cancer medicine |
| 29073095 | Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. | J Matthew Shurtleff | 2017-10-24 | Proceedings of the National Academy of Sciences of the United States of America |
| 27216562 | Genetically engineered pre-microRNA-34a prodrug suppresses orthotopic osteosarcoma xenograft tumor growth via the induction of apoptosis and cell cycle arrest. | Yong Zhao | 2016-05-24 | Scientific reports |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| YBX1-201 | ENST00000321358 | 2979 | 324aa | ENSP00000361626 |
| YBX1-202 | ENST00000332220 | 776 | 216aa | ENSP00000405937 |
| YBX1-203 | ENST00000436427 | 1524 | 374aa | ENSP00000389639 |
| YBX1-204 | ENST00000467957 | 770 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
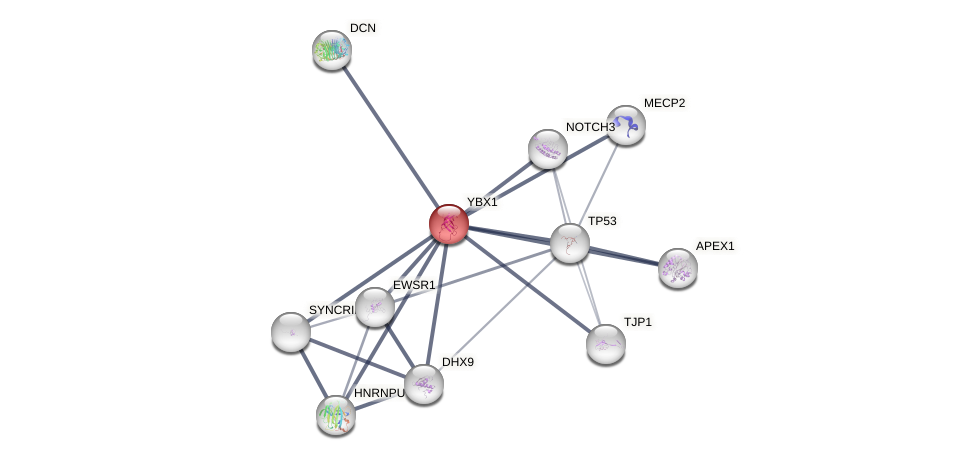
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000065978 | homo_sapiens | ENSP00000007699 | 49.4506 | 40.9341 | homo_sapiens | ENSP00000361626 | 55.5556 | 45.9877 |
| ENSG00000065978 | homo_sapiens | ENSP00000363314 | 50.7177 | 35.8852 | homo_sapiens | ENSP00000361626 | 32.716 | 23.1481 |
| ENSG00000065978 | homo_sapiens | ENSP00000344401 | 45.2 | 33.2 | homo_sapiens | ENSP00000361626 | 34.8765 | 25.6173 |
| ENSG00000065978 | homo_sapiens | ENSP00000228251 | 55.914 | 50 | homo_sapiens | ENSP00000361626 | 64.1975 | 57.4074 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | nomascus_leucogenys | ENSNLEP00000014964 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | pan_paniscus | ENSPPAP00000014831 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | pongo_abelii | ENSPPYP00000001698 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 93.2099 | 91.0494 | pan_troglodytes | ENSPTRP00000078821 | 92.0732 | 89.939 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 31.7901 | 30.5556 | pan_troglodytes | ENSPTRP00000076001 | 82.4 | 79.2 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 93.2099 | 91.0494 | pan_troglodytes | ENSPTRP00000078565 | 92.0732 | 89.939 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 90.1235 | 88.8889 | gorilla_gorilla | ENSGGOP00000024357 | 92.9936 | 91.7197 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 95.679 | 94.4444 | ornithorhynchus_anatinus | ENSOANP00000039174 | 90.1163 | 88.9535 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 88.5802 | 86.4198 | sarcophilus_harrisii | ENSSHAP00000017000 | 87.5 | 85.3659 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 45.0617 | 44.1358 | notamacropus_eugenii | ENSMEUP00000009010 | 59.1093 | 57.8947 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 89.1975 | 88.8889 | dasypus_novemcinctus | ENSDNOP00000017828 | 99.6552 | 99.3103 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | callithrix_jacchus | ENSCJAP00000005817 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | cercocebus_atys | ENSCATP00000004796 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 34.8765 | 32.4074 | macaca_fascicularis | ENSMFAP00000043809 | 90.4 | 84 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | macaca_mulatta | ENSMMUP00000017257 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 83.0247 | 78.7037 | macaca_nemestrina | ENSMNEP00000020831 | 84.5912 | 80.1887 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 80.5556 | 78.3951 | macaca_nemestrina | ENSMNEP00000037428 | 90.625 | 88.1944 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | papio_anubis | ENSPANP00000011056 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 72.8395 | 70.3704 | mandrillus_leucophaeus | ENSMLEP00000032928 | 84.2857 | 81.4286 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 75.3086 | 70.3704 | mandrillus_leucophaeus | ENSMLEP00000027071 | 86.5248 | 80.8511 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 98.7654 | 98.7654 | equus_asinus | ENSEASP00005047744 | 81.4249 | 81.4249 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 99.6914 | 99.3827 | capra_hircus | ENSCHIP00000012408 | 99.6914 | 99.3827 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 95.9877 | 95.3704 | capra_hircus | ENSCHIP00000001450 | 99.6795 | 99.0385 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | panthera_leo | ENSPLOP00000014332 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | bos_indicus_hybrid | ENSBIXP00005001416 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 38.5802 | 38.2716 | oryctolagus_cuniculus | ENSOCUP00000034177 | 98.4252 | 97.6378 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 71.6049 | 69.1358 | equus_caballus | ENSECAP00000034220 | 80.2768 | 77.5087 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 93.2099 | 92.5926 | canis_lupus_familiaris | ENSCAFP00845028940 | 94.9686 | 94.3396 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | canis_lupus_familiaris | ENSCAFP00845022602 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | panthera_pardus | ENSPPRP00000024323 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | balaenoptera_musculus | ENSBMSP00010014258 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 50.9259 | 44.1358 | felis_catus | ENSFCAP00000058166 | 61.5672 | 53.3582 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 68.8272 | 64.5062 | ursus_americanus | ENSUAMP00000023803 | 71.7042 | 67.2026 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 98.7654 | 97.8395 | monodelphis_domestica | ENSMODP00000021480 | 98.7654 | 97.8395 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | monodon_monoceros | ENSMMNP00015010297 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 59.8765 | 55.5556 | sus_scrofa | ENSSSCP00000080329 | 70.5455 | 65.4545 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 91.9753 | 91.0494 | microcebus_murinus | ENSMICP00000041091 | 91.9753 | 91.0494 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 87.3457 | 85.1852 | microcebus_murinus | ENSMICP00000046130 | 89.557 | 87.3418 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 83.9506 | 81.7901 | octodon_degus | ENSODEP00000005422 | 90.3654 | 88.0399 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 74.0741 | 71.6049 | jaculus_jaculus | ENSJJAP00000017743 | 90.566 | 87.5472 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 75.3086 | 72.5309 | jaculus_jaculus | ENSJJAP00000001877 | 86.8327 | 83.6299 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 99.6914 | ovis_aries_rambouillet | ENSOARP00020000272 | 100 | 99.6914 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 90.1235 | 89.8148 | ovis_aries_rambouillet | ENSOARP00020023326 | 65.4709 | 65.2466 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | physeter_catodon | ENSPCTP00005028082 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 30.5556 | 29.321 | aotus_nancymaae | ENSANAP00000033715 | 93.3962 | 89.6226 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 36.7284 | 36.4198 | aotus_nancymaae | ENSANAP00000027669 | 94.4444 | 93.6508 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 33.642 | 33.0247 | aotus_nancymaae | ENSANAP00000035919 | 93.1624 | 91.453 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 78.0864 | 75.6173 | aotus_nancymaae | ENSANAP00000026639 | 89.0845 | 86.2676 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 33.9506 | 32.0988 | saimiri_boliviensis_boliviensis | ENSSBOP00000022949 | 94.0171 | 88.8889 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 99.6914 | 99.6914 | phocoena_sinus | ENSPSNP00000002063 | 99.6914 | 99.6914 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 99.6914 | 99.6914 | phocoena_sinus | ENSPSNP00000015138 | 99.6914 | 99.6914 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 76.2346 | 74.6914 | camelus_dromedarius | ENSCDRP00005011988 | 93.9164 | 92.0152 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 70.9877 | 66.358 | carlito_syrichta | ENSTSYP00000013724 | 87.7863 | 82.0611 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 62.037 | 58.9506 | rattus_norvegicus | ENSRNOP00000079937 | 72.5632 | 68.9531 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 71.6049 | 70.0617 | prolemur_simus | ENSPSMP00000025503 | 88.8889 | 86.9732 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 87.037 | 83.9506 | sorex_araneus | ENSSARP00000011404 | 85.1964 | 82.1752 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 80.5556 | 78.7037 | peromyscus_maniculatus_bairdii | ENSPEMP00000010653 | 93.2143 | 91.0714 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 36.7284 | 35.8025 | peromyscus_maniculatus_bairdii | ENSPEMP00000035987 | 93.7008 | 91.3386 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 36.4198 | 35.1852 | mus_spicilegus | ENSMSIP00000002272 | 95.1613 | 91.9355 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 99.0741 | 98.7654 | cricetulus_griseus_chok1gshd | ENSCGRP00001002360 | 99.6894 | 99.3789 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 29.0123 | 26.5432 | vombatus_ursinus | ENSVURP00010028299 | 78.9916 | 72.2689 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | rhinolophus_ferrumequinum | ENSRFEP00010008720 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | bos_taurus | ENSBTAP00000023094 | 100 | 100 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 71.2963 | 67.284 | rhinopithecus_bieti | ENSRBIP00000021679 | 85.8736 | 81.0409 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 83.9506 | 83.642 | canis_lupus_dingo | ENSCAFP00020032074 | 98.5507 | 98.1884 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 100 | 100 | canis_lupus_dingo | ENSCAFP00020030884 | 99.0826 | 99.0826 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 98.7654 | 97.8395 | phascolarctos_cinereus | ENSPCIP00000011392 | 98.7654 | 97.8395 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 70.9877 | 67.5926 | nannospalax_galili | ENSNGAP00000001517 | 87.4525 | 83.27 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 89.5062 | 89.5062 | cervus_hanglu_yarkandensis | ENSCHYP00000031451 | 85.5457 | 85.5457 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 64.1975 | 60.1852 | otolemur_garnettii | ENSOGAP00000016311 | 75.6364 | 70.9091 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 57.716 | 54.321 | otolemur_garnettii | ENSOGAP00000017914 | 85.7798 | 80.7339 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 85.1852 | 82.716 | cebus_imitator | ENSCCAP00000013666 | 88.1789 | 85.623 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 78.3951 | 75.9259 | moschus_moschiferus | ENSMMSP00000022532 | 84.3854 | 81.7276 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 71.9136 | 67.9012 | rhinopithecus_roxellana | ENSRROP00000011675 | 86.2963 | 81.4815 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 68.8272 | 65.4321 | rhinopithecus_roxellana | ENSRROP00000009768 | 85.7692 | 81.5385 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 32.0988 | 32.0988 | propithecus_coquereli | ENSPCOP00000006358 | 83.871 | 83.871 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 41.0494 | 31.1728 | caenorhabditis_elegans | Y39A1C.3.1 | 45.2381 | 34.3537 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 34.8765 | 24.6914 | caenorhabditis_elegans | F46F11.2.1 | 42.3221 | 29.9625 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 37.3457 | 25.6173 | caenorhabditis_elegans | M01E11.5.1 | 45.6604 | 31.3208 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 25.3086 | 16.6667 | drosophila_melanogaster | FBpp0076798 | 42.0513 | 27.6923 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 24.3827 | 15.7407 | ciona_intestinalis | ENSCINP00000032715 | 40.7216 | 26.2887 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 44.7531 | 40.7407 | eptatretus_burgeri | ENSEBUP00000011763 | 39.0836 | 35.5795 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 73.7654 | 66.0494 | callorhinchus_milii | ENSCMIP00000017079 | 78.3607 | 70.1639 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 81.4815 | 76.2346 | latimeria_chalumnae | ENSLACP00000020639 | 86.5574 | 80.9836 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 79.9383 | 73.7654 | lepisosteus_oculatus | ENSLOCP00000002985 | 83.2797 | 76.8489 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 79.321 | 70.9877 | danio_rerio | ENSDARP00000010482 | 83.1715 | 74.4337 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 79.321 | 71.2963 | carassius_auratus | ENSCARP00000052571 | 82.3718 | 74.0385 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 77.4691 | 69.1358 | carassius_auratus | ENSCARP00000083868 | 78.1931 | 69.7819 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 79.321 | 71.6049 | astyanax_mexicanus | ENSAMXP00000014898 | 82.3718 | 74.359 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 78.0864 | 71.6049 | ictalurus_punctatus | ENSIPUP00000032433 | 81.3505 | 74.5981 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 70.9877 | 64.5062 | esox_lucius | ENSELUP00000005321 | 75.6579 | 68.75 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 77.7778 | 70.0617 | electrophorus_electricus | ENSEEEP00000041050 | 80.7692 | 72.7564 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 69.7531 | 62.3457 | oncorhynchus_kisutch | ENSOKIP00005046670 | 63.1285 | 56.4246 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 70.3704 | 63.5802 | oncorhynchus_kisutch | ENSOKIP00005069427 | 74.7541 | 67.541 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 70.679 | 62.037 | oncorhynchus_mykiss | ENSOMYP00000037935 | 72.0126 | 63.2075 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 75 | 66.358 | oncorhynchus_mykiss | ENSOMYP00000046087 | 75.7009 | 66.9782 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 59.5679 | 51.8519 | salmo_salar | ENSSSAP00000015202 | 69.4245 | 60.4317 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 75.6173 | 68.2099 | salmo_salar | ENSSSAP00000072764 | 75.1534 | 67.7914 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 75.6173 | 68.2099 | salmo_trutta | ENSSTUP00000099220 | 76.087 | 68.6335 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 74.6914 | 66.358 | salmo_trutta | ENSSTUP00000038088 | 75.3894 | 66.9782 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 62.963 | 54.9383 | gadus_morhua | ENSGMOP00000031443 | 61.6314 | 53.7764 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 71.6049 | 66.0494 | fundulus_heteroclitus | ENSFHEP00000001325 | 76.5677 | 70.6271 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 73.1481 | 66.6667 | poecilia_reticulata | ENSPREP00000025294 | 79.2642 | 72.2408 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 73.1481 | 66.6667 | xiphophorus_maculatus | ENSXMAP00000018726 | 78.2178 | 71.2871 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 72.8395 | 66.358 | oryzias_latipes | ENSORLP00000005291 | 77.1242 | 70.2614 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 70.9877 | 64.8148 | cyclopterus_lumpus | ENSCLMP00005005870 | 72.327 | 66.0377 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 68.8272 | 61.4198 | oreochromis_niloticus | ENSONIP00000037722 | 73.1148 | 65.2459 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 72.5309 | 66.358 | haplochromis_burtoni | ENSHBUP00000008545 | 77.3026 | 70.7237 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 78.0864 | 70.679 | astatotilapia_calliptera | ENSACLP00000033494 | 78.8162 | 71.3396 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 70.9877 | 63.5802 | sparus_aurata | ENSSAUP00010001429 | 75.9076 | 67.9868 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 40.7407 | 36.1111 | sparus_aurata | ENSSAUP00010062302 | 62.2642 | 55.1887 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 78.0864 | 70.9877 | lates_calcarifer | ENSLCAP00010059078 | 78.5714 | 71.4286 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 86.1111 | 80.8642 | xenopus_tropicalis | ENSXETP00000029443 | 87.1875 | 81.875 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 55.2469 | 46.2963 | chrysemys_picta_bellii | ENSCPBP00000002056 | 59.6667 | 50 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 95.3704 | 91.6667 | sphenodon_punctatus | ENSSPUP00000012006 | 91.1504 | 87.6106 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 52.1605 | 39.8148 | crocodylus_porosus | ENSCPRP00005003533 | 55.5921 | 42.4342 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 92.5926 | 88.5802 | laticauda_laticaudata | ENSLLTP00000006086 | 86.7052 | 82.948 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 92.5926 | 88.5802 | notechis_scutatus | ENSNSUP00000025976 | 86.7052 | 82.948 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 92.5926 | 88.5802 | pseudonaja_textilis | ENSPTXP00000022632 | 86.7052 | 82.948 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 94.4444 | 91.358 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000030195 | 79.0698 | 76.4858 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 91.9753 | 89.1975 | gallus_gallus | ENSGALP00010025081 | 90.8537 | 88.1098 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 80.5556 | 77.7778 | meleagris_gallopavo | ENSMGAP00000007005 | 81.8182 | 78.9969 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 75.9259 | 72.5309 | serinus_canaria | ENSSCAP00000022714 | 96.0938 | 91.7969 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 95.0617 | 91.6667 | parus_major | ENSPMJP00000003478 | 96.25 | 92.8125 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 72.5309 | 66.358 | neolamprologus_brichardi | ENSNBRP00000003989 | 78.3333 | 71.6667 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 94.4444 | 91.358 | podarcis_muralis | ENSPMRP00000031645 | 85.7143 | 82.9132 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 38.2716 | 36.4198 | geospiza_fortis | ENSGFOP00000015224 | 98.4127 | 93.6508 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 95.0617 | 91.6667 | taeniopygia_guttata | ENSTGUP00000017543 | 95.0617 | 91.6667 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 80.2469 | 74.6914 | erpetoichthys_calabaricus | ENSECRP00000016067 | 82.5397 | 76.8254 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 73.4568 | 66.0494 | kryptolebias_marmoratus | ENSKMAP00000017481 | 78.5479 | 70.6271 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 93.5185 | 90.7407 | salvator_merianae | ENSSMRP00000009928 | 87.5723 | 84.9711 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 78.0864 | 70.0617 | oryzias_javanicus | ENSOJAP00000035792 | 77.6074 | 69.6319 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 67.284 | 57.0988 | amphilophus_citrinellus | ENSACIP00000028970 | 67.7019 | 57.4534 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 76.8519 | 69.7531 | dicentrarchus_labrax | ENSDLAP00005015119 | 76.3804 | 69.3252 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 69.7531 | 63.5802 | amphiprion_percula | ENSAPEP00000020476 | 73.6156 | 67.101 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 73.1481 | 66.358 | oryzias_sinensis | ENSOSIP00000017981 | 77.1987 | 70.0326 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 45.0617 | 37.6543 | pelodiscus_sinensis | ENSPSIP00000010512 | 66.0633 | 55.2036 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 50 | 41.0494 | gopherus_evgoodei | ENSGEVP00005016914 | 60.4478 | 49.6269 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 52.7778 | 42.9012 | chelonoidis_abingdonii | ENSCABP00000009378 | 58.9655 | 47.931 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 72.8395 | 67.284 | seriola_dumerili | ENSSDUP00000028917 | 78.1457 | 72.1854 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 70.3704 | 63.8889 | cottoperca_gobio | ENSCGOP00000001748 | 76.2542 | 69.2308 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 85.4938 | 82.4074 | struthio_camelus_australis | ENSSCUP00000006245 | 95.8477 | 92.3875 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 89.5062 | 86.4198 | anser_brachyrhynchus | ENSABRP00000006914 | 96.0265 | 92.7152 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 71.6049 | 65.4321 | gasterosteus_aculeatus | ENSGACP00000008523 | 76.3158 | 69.7368 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 63.8889 | 56.4815 | cynoglossus_semilaevis | ENSCSEP00000030081 | 68.543 | 60.596 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 78.3951 | 70.679 | poecilia_formosa | ENSPFOP00000011455 | 79.1277 | 71.3396 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 79.9383 | 71.6049 | myripristis_murdjan | ENSMMDP00005020502 | 80.1858 | 71.8266 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 77.4691 | 69.7531 | sander_lucioperca | ENSSLUP00000019812 | 77.4691 | 69.7531 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 95.9877 | 92.9012 | strigops_habroptila | ENSSHBP00005001009 | 96.2848 | 93.1889 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 77.7778 | 70.9877 | amphiprion_ocellaris | ENSAOCP00000023212 | 78.2609 | 71.4286 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 53.7037 | 43.2099 | terrapene_carolina_triunguis | ENSTMTP00000010807 | 60.4167 | 48.6111 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 69.7531 | 62.3457 | hucho_hucho | ENSHHUP00000045085 | 75.3333 | 67.3333 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 75.6173 | 67.9012 | hucho_hucho | ENSHHUP00000060722 | 76.087 | 68.323 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 73.7654 | 66.9753 | betta_splendens | ENSBSLP00000005996 | 75.873 | 68.8889 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 79.0123 | 72.2222 | pygocentrus_nattereri | ENSPNAP00000021684 | 83.1169 | 75.974 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 77.1605 | 70.679 | anabas_testudineus | ENSATEP00000018037 | 77.8816 | 71.3396 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 21.2963 | 12.963 | ciona_savignyi | ENSCSAVP00000011162 | 39.4286 | 24 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 72.8395 | 67.284 | seriola_lalandi_dorsalis | ENSSLDP00000031704 | 78.1457 | 72.1854 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 71.9136 | 65.7407 | labrus_bergylta | ENSLBEP00000007126 | 76.8977 | 70.297 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 72.5309 | 66.358 | pundamilia_nyererei | ENSPNYP00000013294 | 78.3333 | 71.6667 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 78.3951 | 71.2963 | mastacembelus_armatus | ENSMAMP00000027156 | 78.882 | 71.7391 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 78.0864 | 70.679 | stegastes_partitus | ENSSPAP00000027434 | 78.0864 | 70.679 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 75 | 66.358 | oncorhynchus_tshawytscha | ENSOTSP00005083434 | 64.1161 | 56.7282 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 70.3704 | 63.8889 | oncorhynchus_tshawytscha | ENSOTSP00005004916 | 75.7475 | 68.7708 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 75 | 66.358 | oncorhynchus_tshawytscha | ENSOTSP00005055525 | 64.1161 | 56.7282 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 72.8395 | 66.9753 | acanthochromis_polyacanthus | ENSAPOP00000015358 | 77.3771 | 71.1475 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 95.3704 | 92.5926 | aquila_chrysaetos_chrysaetos | ENSACCP00020006867 | 96.2617 | 93.4579 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 74.0741 | 66.9753 | nothobranchius_furzeri | ENSNFUP00015050735 | 75.7098 | 68.4543 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 72.2222 | 65.7407 | cyprinodon_variegatus | ENSCVAP00000020736 | 78.2609 | 71.2375 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 61.1111 | 55.8642 | denticeps_clupeoides | ENSDCDP00000018221 | 77.9528 | 71.2598 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 63.8889 | 57.0988 | denticeps_clupeoides | ENSDCDP00000021312 | 76.6667 | 68.5185 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 58.642 | 53.7037 | denticeps_clupeoides | ENSDCDP00000016350 | 76.6129 | 70.1613 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 41.6667 | 38.5802 | denticeps_clupeoides | ENSDCDP00000013328 | 71.0526 | 65.7895 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 58.9506 | 50.3086 | denticeps_clupeoides | ENSDCDP00000027158 | 60.0629 | 51.2579 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 66.0494 | 59.8765 | denticeps_clupeoides | ENSDCDP00000012188 | 74.0484 | 67.128 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 41.0494 | 38.2716 | denticeps_clupeoides | ENSDCDP00000012754 | 64.8781 | 60.4878 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 37.963 | 36.1111 | denticeps_clupeoides | ENSDCDP00000020333 | 62.7551 | 59.6939 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 66.9753 | 59.8765 | denticeps_clupeoides | ENSDCDP00000012069 | 74.5704 | 66.6667 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 40.1235 | 36.7284 | denticeps_clupeoides | ENSDCDP00000001561 | 63.1068 | 57.767 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 64.5062 | 58.3333 | denticeps_clupeoides | ENSDCDP00000015027 | 69.4352 | 62.7907 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 67.5926 | 60.8025 | denticeps_clupeoides | ENSDCDP00000014876 | 74.2373 | 66.7797 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 59.8765 | 54.0123 | denticeps_clupeoides | ENSDCDP00000051093 | 75.4864 | 68.0934 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 66.358 | 60.1852 | denticeps_clupeoides | ENSDCDP00000015237 | 73.8832 | 67.0103 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 78.0864 | 71.6049 | denticeps_clupeoides | ENSDCDP00000045578 | 82.1429 | 75.3247 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 64.8148 | 58.3333 | denticeps_clupeoides | ENSDCDP00000013515 | 74.4681 | 67.0213 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 58.0247 | 53.0864 | denticeps_clupeoides | ENSDCDP00000016360 | 75.2 | 68.8 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 43.2099 | 35.1852 | denticeps_clupeoides | ENSDCDP00000000379 | 54.4747 | 44.358 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 66.9753 | 59.8765 | denticeps_clupeoides | ENSDCDP00000058781 | 74.5704 | 66.6667 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 63.8889 | 57.0988 | denticeps_clupeoides | ENSDCDP00000011825 | 76.6667 | 68.5185 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 67.5926 | 61.4198 | denticeps_clupeoides | ENSDCDP00000002895 | 75.2577 | 68.3849 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 40.4321 | 37.963 | denticeps_clupeoides | ENSDCDP00000001365 | 63.9024 | 60 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 61.1111 | 54.9383 | denticeps_clupeoides | ENSDCDP00000036108 | 69.2308 | 62.2378 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 55.5556 | 47.5309 | denticeps_clupeoides | ENSDCDP00000021069 | 55.5556 | 47.5309 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 55.2469 | 47.2222 | denticeps_clupeoides | ENSDCDP00000013618 | 56.2893 | 48.1132 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 66.9753 | 60.1852 | denticeps_clupeoides | ENSDCDP00000011059 | 73.5593 | 66.1017 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 41.0494 | 35.8025 | denticeps_clupeoides | ENSDCDP00000054010 | 72.6776 | 63.388 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 77.4691 | 70.3704 | larimichthys_crocea | ENSLCRP00005051231 | 77.9503 | 70.8075 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 71.2963 | 63.8889 | takifugu_rubripes | ENSTRUP00000086103 | 71.0769 | 63.6923 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 76.5432 | 72.5309 | ficedula_albicollis | ENSFALP00000010756 | 89.2086 | 84.5324 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 70.0617 | 64.5062 | hippocampus_comes | ENSHCOP00000014044 | 74.9175 | 68.9769 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 79.6296 | 70.9877 | cyprinus_carpio_carpio | ENSCCRP00000056047 | 82.1656 | 73.2484 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 78.7037 | 70.679 | cyprinus_carpio_carpio | ENSCCRP00000002346 | 82.7922 | 74.3506 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 96.2963 | 92.5926 | coturnix_japonica | ENSCJPP00005007794 | 95.7055 | 92.0245 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 73.1481 | 66.6667 | poecilia_latipinna | ENSPLAP00000028355 | 78.2178 | 71.2871 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 70.3704 | 62.3457 | sinocyclocheilus_grahami | ENSSGRP00000060742 | 71.4734 | 63.3229 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 79.9383 | 71.6049 | sinocyclocheilus_grahami | ENSSGRP00000016743 | 80.6854 | 72.2741 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 72.5309 | 66.358 | maylandia_zebra | ENSMZEP00005027413 | 78.3333 | 71.6667 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 70.3704 | 64.1975 | tetraodon_nigroviridis | ENSTNIP00000014949 | 76.5101 | 69.7987 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 69.7531 | 62.6543 | paramormyrops_kingsleyae | ENSPKIP00000033287 | 59.7884 | 53.7037 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 77.1605 | 69.7531 | paramormyrops_kingsleyae | ENSPKIP00000032363 | 62.3441 | 56.3591 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 39.8148 | 31.7901 | scleropages_formosus | ENSSFOP00015047455 | 57.5893 | 45.9821 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 29.0123 | 26.2346 | leptobrachium_leishanense | ENSLLEP00000011948 | 87.8505 | 79.4393 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 53.7037 | 49.3827 | leptobrachium_leishanense | ENSLLEP00000041487 | 74.359 | 68.3761 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 82.4074 | 75.9259 | leptobrachium_leishanense | ENSLLEP00000018507 | 72.3577 | 66.6667 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 75.6173 | 70.3704 | leptobrachium_leishanense | ENSLLEP00000005650 | 88.4477 | 82.3105 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 72.2222 | 66.358 | scophthalmus_maximus | ENSSMAP00000018545 | 77.2277 | 70.9571 |
| ENSG00000065978 | homo_sapiens | ENSP00000361626 | 73.1481 | 66.358 | oryzias_melastigma | ENSOMEP00000024267 | 77.7049 | 70.4918 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0003676 | enables nucleic acid binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0003677 | enables DNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0003690 | enables double-stranded DNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 2 | HDA,IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005576 | located_in extracellular region | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005634 | is_active_in nucleus | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005689 | part_of U12-type spliceosomal complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005783 | located_in endoplasmic reticulum | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005886 | located_in plasma membrane | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0006355 | involved_in regulation of DNA-templated transcription | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006397 | involved_in mRNA processing | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008380 | involved_in RNA splicing | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008544 | involved_in epidermis development | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0010468 | involved_in regulation of gene expression | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0010494 | located_in cytoplasmic stress granule | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0017148 | involved_in negative regulation of translation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0035198 | enables miRNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0043231 | located_in intracellular membrane-bounded organelle | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0048255 | involved_in mRNA stabilization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0048598 | involved_in embryonic morphogenesis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0050658 | involved_in RNA transport | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0051020 | enables GTPase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0051031 | involved_in tRNA transport | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0051781 | involved_in positive regulation of cell division | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0062153 | enables C5-methylcytidine-containing RNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0070062 | located_in extracellular exosome | 2 | HDA,IDA | Homo_sapiens(9606) | Component |
| GO:0070934 | involved_in CRD-mediated mRNA stabilization | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0070937 | part_of CRD-mediated mRNA stability complex | 2 | IDA,IPI | Homo_sapiens(9606) | Component |
| GO:0071204 | part_of histone pre-mRNA 3end processing complex | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:1900152 | involved_in negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1903608 | involved_in protein localization to cytoplasmic stress granule | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1990124 | part_of messenger ribonucleoprotein complex | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:1990428 | involved_in miRNA transport | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1990837 | enables sequence-specific double-stranded DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:1990904 | part_of ribonucleoprotein complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:2000767 | involved_in positive regulation of cytoplasmic translation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:2000773 | involved_in negative regulation of cellular senescence | 1 | ISS | Homo_sapiens(9606) | Process |