RBP Type
Non-canonical_RBPs
Diseases
Drug
Main interacting RNAs
N.A.
Moonlighting functions
Kinase
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000065883 Gene ID
8621 Accession
1733
Symbol
CDK13
Alias
CHED;CDC2L;CDC2L5;hCDK13;CHDFIDD
Full Name
cyclin dependent kinase 13
Status
Confidence
Length
149460 bases
Strand
Plus strand
Position
7 : 39950121 - 40099580
RNA binding domain
N.A.
Summary
The protein encoded by this gene is a member of the cyclin-dependent serine/threonine protein kinase family. Members of this family are well known for their essential roles as master switches in cell cycle control. The exact function of this protein has not yet been determined, but it may play a role in mRNA processing and may be involved in regulation of hematopoiesis. Alternatively spliced transcript variants have been described.[provided by RefSeq, Dec 2009]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
30409083 Therapeutic targeting of transcriptional cyclin-dependent kinases.
D Matthew Galbraith
2019-04-01
Transcription
31652541 CDK12 Activity-Dependent Phosphorylation Events in Human Cells.
Bartlomiej Bartkowiak
2019-10-22
Biomolecules
25561469 Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing.
Kaiwei Liang
2015-03-01
Molecular and cellular biology
26886422 CDK13, a Kinase Involved in Pre-mRNA Splicing, Is a Component of the Perinucleolar Compartment.
Yasmine Even
2016-01-01
PloS one
30988284 CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation.
Malgorzata Krajewska
2019-04-15
Nature communications
27571479 Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors.
Tinghu Zhang
2016-10-01
Nature chemical biology
Name
Transcript ID
bp
Protein
Translation ID
CDK13-202
ENST00000340829
5455
1452aa
ENSP00000340557
CDK13-201
ENST00000181839
9525
1512aa
ENSP00000181839
CDK13-211
ENST00000643868
1901
279aa
ENSP00000495083
CDK13-217
ENST00000646039
4466
1308aa
ENSP00000494168
CDK13-210
ENST00000643859
3838
1099aa
ENSP00000496440
CDK13-220
ENST00000647518
4190
No protein
-
CDK13-208
ENST00000642626
1150
288aa
ENSP00000493853
CDK13-209
ENST00000642660
948
No protein
-
CDK13-218
ENST00000646437
772
257aa
ENSP00000496618
CDK13-205
ENST00000484589
5166
456aa
ENSP00000494206
CDK13-212
ENST00000643915
3276
915aa
ENSP00000496187
CDK13-219
ENST00000647453
2998
No protein
-
CDK13-206
ENST00000642213
1053
No protein
-
CDK13-215
ENST00000645470
900
239aa
ENSP00000495036
CDK13-221
ENST00000700485
2672
No protein
-
CDK13-222
ENST00000700486
2901
No protein
-
CDK13-223
ENST00000700487
2858
No protein
-
CDK13-207
ENST00000642592
1047
143aa
ENSP00000494207
CDK13-204
ENST00000478563
904
No protein
-
CDK13-213
ENST00000644221
4511
No protein
-
CDK13-214
ENST00000644561
2576
No protein
-
CDK13-216
ENST00000645826
2250
No protein
-
CDK13-203
ENST00000465643
2394
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
37227 Platelet Function Tests 4.3500000E-006 -
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
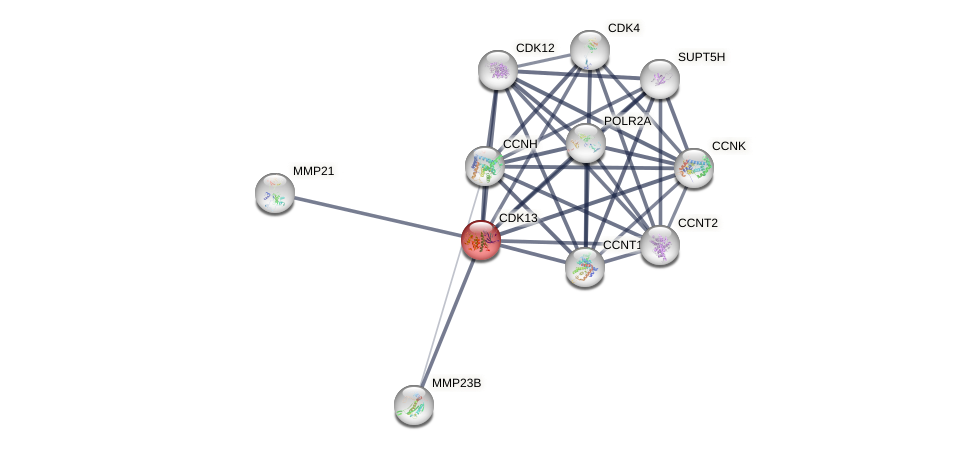
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000380 involved_in alternative mRNA splicing, via spliceosome 2 ISS,TAS Homo_sapiens(9606) Process GO:0002945 part_of cyclin K-CDK13 complex 1 IPI Homo_sapiens(9606) Component GO:0003723 enables RNA binding 1 HDA Homo_sapiens(9606) Function GO:0004672 enables protein kinase activity 1 TAS Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005524 enables ATP binding 1 IEA Homo_sapiens(9606) Function GO:0005576 located_in extracellular region 1 TAS Homo_sapiens(9606) Component GO:0005615 located_in extracellular space 1 HDA Homo_sapiens(9606) Component GO:0005634 is_active_in nucleus 1 IBA Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 1 TAS Homo_sapiens(9606) Component GO:0005794 located_in Golgi apparatus 1 IDA Homo_sapiens(9606) Component GO:0005829 located_in cytosol 1 IDA Homo_sapiens(9606) Component GO:0006468 involved_in protein phosphorylation 1 IBA Homo_sapiens(9606) Process GO:0008024 part_of cyclin/CDK positive transcription elongation factor complex 1 IBA Homo_sapiens(9606) Component GO:0008353 enables RNA polymerase II CTD heptapeptide repeat kinase activity 2 IBA,IDA Homo_sapiens(9606) Function GO:0009966 involved_in regulation of signal transduction 1 IDA Homo_sapiens(9606) Process GO:0016607 located_in nuclear speck 1 IDA Homo_sapiens(9606) Component GO:0019901 enables protein kinase binding 1 IEA Homo_sapiens(9606) Function GO:0019908 part_of nuclear cyclin-dependent protein kinase holoenzyme complex 1 ISS Homo_sapiens(9606) Component GO:0030097 involved_in hemopoiesis 1 IMP Homo_sapiens(9606) Process GO:0030332 enables cyclin binding 2 IBA,IPI Homo_sapiens(9606) Function GO:0032968 involved_in positive regulation of transcription elongation by RNA polymerase II 2 IBA,IDA Homo_sapiens(9606) Process GO:0045944 involved_in positive regulation of transcription by RNA polymerase II 1 IDA Homo_sapiens(9606) Process GO:0051726 involved_in regulation of cell cycle 1 IEA Homo_sapiens(9606) Process GO:0106310 enables protein serine kinase activity 1 IEA Homo_sapiens(9606) Function GO:1904813 located_in ficolin-1-rich granule lumen 1 TAS Homo_sapiens(9606) Component GO:2000737 involved_in negative regulation of stem cell differentiation 1 IEA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.