Welcome to
RBP World!
Functions and Diseases
| RBP Type | Non-canonical_RBPs |
| Diseases | Disease |
| Drug | N.A. |
| Main interacting RNAs | N.A. |
| Moonlighting functions | N.A. |
| Localizations | Stress granucleNucleoli |
| BulkPerturb-seq |
Description
| Ensembl ID | ENSG00000013573 | Gene ID | 1663 | Accession | 2736 |
| Symbol | DDX11 | Alias | CHL1;KRG2;WABS;CHLR1 | Full Name | DEAD/H-box helicase 11 |
| Status | Confidence | Length | 30940 bases | Strand | Plus strand |
| Position | 12 : 31073860 - 31104799 | RNA binding domain | N.A. | ||
| Summary | DEAD box proteins, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD), are putative RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure such as translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. Based on their distribution patterns, some members of this family are believed to be involved in embryogenesis, spermatogenesis, and cellular growth and division. This gene encodes a DEAD box protein, which is an enzyme that possesses both ATPase and DNA helicase activities. This gene is a homolog of the yeast CHL1 gene, and may function to maintain chromosome transmission fidelity and genome stability. Alternative splicing results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 30469382 | Molecular and Cellular Functions of the Warsaw Breakage Syndrome DNA Helicase DDX11. | M Francesca Pisani | 2018-11-21 | Genes |
| 33669056 | Role of the DDX11 DNA Helicase in Warsaw Breakage Syndrome Etiology. | Diana Santos | 2021-02-25 | International journal of molecular sciences |
| 31824187 | Spotlight on Warsaw Breakage Syndrome. | M Francesca Pisani | 2019-01-01 | The application of clinical genetics |
| 34281459 | Long non-coding RNA DDX11-AS1 promotes esophageal carcinoma cell proliferation and migration through regulating the miR-514b-3p/RBX1 axis. | Chao Wu | 2021-12-01 | Bioengineered |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| DDX11-218 | ENST00000542129 | 1561 | 304aa | ENSP00000442466 |
| DDX11-201 | ENST00000228264 | 3717 | 880aa | ENSP00000228264 |
| DDX11-223 | ENST00000542838 | 3920 | 906aa | ENSP00000443426 |
| DDX11-205 | ENST00000438391 | 1135 | 302aa | ENSP00000407646 |
| DDX11-204 | ENST00000435753 | 4182 | 288aa | ENSP00000406799 |
| DDX11-217 | ENST00000540935 | 1210 | 288aa | ENSP00000443578 |
| DDX11-213 | ENST00000539049 | 3471 | 297aa | ENSP00000445568 |
| DDX11-208 | ENST00000536265 | 3603 | No protein | - |
| DDX11-203 | ENST00000415475 | 883 | 224aa | ENSP00000406457 |
| DDX11-229 | ENST00000545668 | 3751 | 970aa | ENSP00000440402 |
| DDX11-202 | ENST00000350437 | 3724 | 856aa | ENSP00000309965 |
| DDX11-206 | ENST00000535158 | 1718 | No protein | - |
| DDX11-226 | ENST00000543756 | 613 | 78aa | ENSP00000439298 |
| DDX11-220 | ENST00000542244 | 1193 | 171aa | ENSP00000441425 |
| DDX11-207 | ENST00000535317 | 879 | 191aa | ENSP00000440171 |
| DDX11-227 | ENST00000544652 | 400 | 91aa | ENSP00000473610 |
| DDX11-211 | ENST00000538345 | 498 | 31aa | ENSP00000445219 |
| DDX11-219 | ENST00000542242 | 795 | No protein | - |
| DDX11-224 | ENST00000543026 | 2066 | No protein | - |
| DDX11-230 | ENST00000545717 | 617 | No protein | - |
| DDX11-209 | ENST00000536580 | 838 | No protein | - |
| DDX11-214 | ENST00000539673 | 652 | No protein | - |
| DDX11-225 | ENST00000543511 | 571 | No protein | - |
| DDX11-215 | ENST00000539699 | 569 | No protein | - |
| DDX11-221 | ENST00000542661 | 562 | No protein | - |
| DDX11-212 | ENST00000538740 | 750 | No protein | - |
| DDX11-222 | ENST00000542777 | 489 | No protein | - |
| DDX11-210 | ENST00000537136 | 482 | No protein | - |
| DDX11-228 | ENST00000545115 | 545 | 76aa | ENSP00000443750 |
| DDX11-216 | ENST00000539702 | 951 | 172aa | ENSP00000441015 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 49342 | Child Development Disorders, Pervasive | 1.6160000E-005 | - |
| 49345 | Child Development Disorders, Pervasive | 2.2221000E-005 | - |
| 49349 | Child Development Disorders, Pervasive | 2.2221000E-005 | - |
| 49354 | Child Development Disorders, Pervasive | 2.2221000E-005 | - |
| 50070 | Child Development Disorders, Pervasive | 4.0257000E-006 | - |
| 50073 | Child Development Disorders, Pervasive | 5.8312000E-006 | - |
| 50078 | Child Development Disorders, Pervasive | 5.8312000E-006 | - |
| 50081 | Child Development Disorders, Pervasive | 5.8312000E-006 | - |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
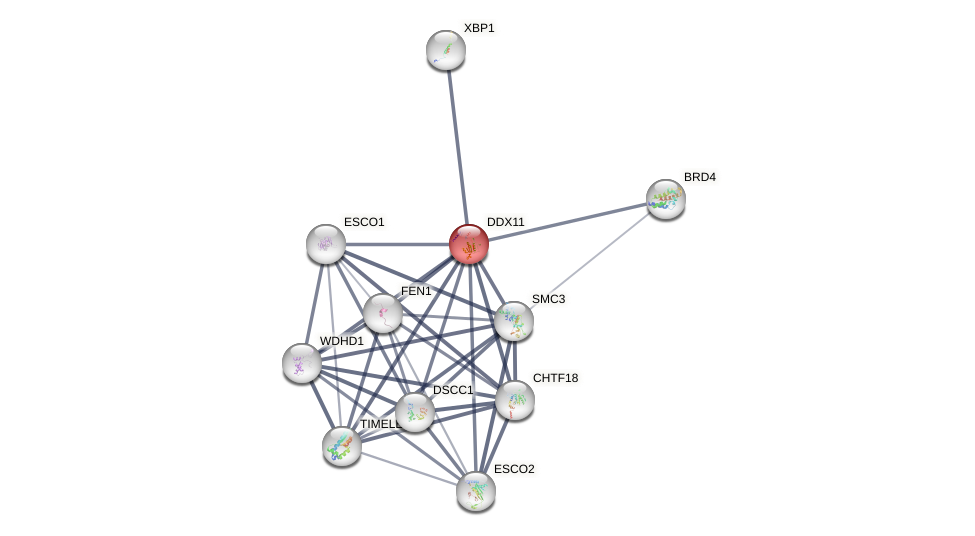
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000785 | part_of chromatin | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0000922 | located_in spindle pole | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0003677 | enables DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003678 | enables DNA helicase activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0003682 | enables chromatin binding | 2 | IDA,IMP | Homo_sapiens(9606) | Function |
| GO:0003688 | enables DNA replication origin binding | 1 | IMP | Homo_sapiens(9606) | Function |
| GO:0003690 | enables double-stranded DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003697 | enables single-stranded DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003727 | enables single-stranded RNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0004386 | enables helicase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005524 | enables ATP binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005634 | colocalizes_with nucleus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005730 | located_in nucleolus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005813 | located_in centrosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0006281 | involved_in DNA repair | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006974 | involved_in DNA damage response | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0007062 | involved_in sister chromatid cohesion | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0008094 | enables ATP-dependent activity, acting on DNA | 2 | IDA,IMP | Homo_sapiens(9606) | Function |
| GO:0008186 | enables ATP-dependent activity, acting on RNA | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0016887 | enables ATP hydrolysis activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0030496 | located_in midbody | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0031297 | involved_in replication fork processing | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032079 | involved_in positive regulation of endodeoxyribonuclease activity | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032091 | involved_in negative regulation of protein binding | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032508 | involved_in DNA duplex unwinding | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0034085 | involved_in establishment of sister chromatid cohesion | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0035563 | involved_in positive regulation of chromatin binding | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0043139 | enables 5-3 DNA helicase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0044806 | involved_in G-quadruplex DNA unwinding | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045142 | enables triplex DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0045876 | involved_in positive regulation of sister chromatid cohesion | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0046872 | enables metal ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0051539 | enables 4 iron, 4 sulfur cluster binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0051880 | enables G-quadruplex DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0070062 | located_in extracellular exosome | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0072711 | involved_in cellular response to hydroxyurea | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0072719 | involved_in cellular response to cisplatin | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1901838 | involved_in positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1904976 | involved_in cellular response to bleomycin | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1990700 | involved_in nucleolar chromatin organization | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:2000781 | involved_in positive regulation of double-strand break repair | 1 | IMP | Homo_sapiens(9606) | Process |