RBP Type
Non-canonical_RBPs
Diseases
N.A.
Drug
Main interacting RNAs
N.A.
Moonlighting functions
N.A.
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000011485 Gene ID
5536 Accession
9322
Symbol
PPP5C
Alias
PP5;PPT;PPP5
Full Name
protein phosphatase 5 catalytic subunit
Status
Confidence
Length
45895 bases
Strand
Plus strand
Position
19 : 46347087 - 46392981
RNA binding domain
N.A.
Summary
This gene encodes a serine/threonine phosphatase which is a member of the protein phosphatase catalytic subunit family. Proteins in this family participate in pathways regulated by reversible phosphorylation at serine and threonine residues; many of these pathways are involved in the regulation of cell growth and differentiation. The product of this gene has been shown to participate in signaling pathways in response to hormones or cellular stress, and elevated levels of this protein may be associated with breast cancer development. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2011]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
PPP5C-201
ENST00000012443
2139
499aa
ENSP00000012443
PPP5C-205
ENST00000478046
1886
485aa
ENSP00000434329
PPP5C-202
ENST00000391919
1929
371aa
ENSP00000375786
PPP5C-204
ENST00000467902
399
No protein
-
PPP5C-212
ENST00000527193
548
No protein
-
PPP5C-207
ENST00000487483
4675
No protein
-
PPP5C-214
ENST00000532058
1970
No protein
-
PPP5C-206
ENST00000486994
2209
No protein
-
PPP5C-203
ENST00000467502
1235
No protein
-
PPP5C-210
ENST00000493347
431
No protein
-
PPP5C-215
ENST00000595055
556
No protein
-
PPP5C-208
ENST00000491003
878
No protein
-
PPP5C-209
ENST00000492109
848
No protein
-
PPP5C-211
ENST00000525507
1300
No protein
-
PPP5C-213
ENST00000527623
1215
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
61721 Crohn Disease 9E-12 26192919 62824 Inflammatory Bowel Diseases 2E-8 26192919 64520 Crohn Disease 4E-7 28067908 96826 Crohn Disease 2E-10 23128233
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
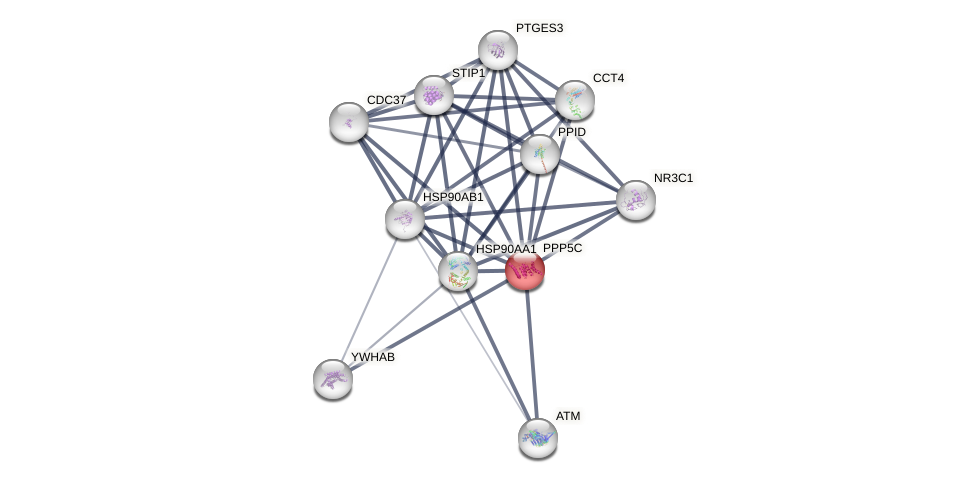
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000165 involved_in MAPK cascade 1 TAS Homo_sapiens(9606) Process GO:0000278 involved_in mitotic cell cycle 1 TAS Homo_sapiens(9606) Process GO:0003723 enables RNA binding 1 IEA Homo_sapiens(9606) Function GO:0004721 enables phosphoprotein phosphatase activity 2 ISS,TAS Homo_sapiens(9606) Function GO:0004722 enables protein serine/threonine phosphatase activity 4 IBA,IDA,ISS,TAS Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005524 enables ATP binding 1 IDA Homo_sapiens(9606) Function GO:0005634 is_active_in nucleus 1 IBA Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 1 TAS Homo_sapiens(9606) Component GO:0005829 is_active_in cytosol 3 IBA,IDA,TAS Homo_sapiens(9606) Component GO:0005886 located_in plasma membrane 1 IEA Homo_sapiens(9606) Component GO:0006302 involved_in double-strand break repair 1 TAS Homo_sapiens(9606) Process GO:0006351 involved_in DNA-templated transcription 1 TAS Homo_sapiens(9606) Process GO:0006470 involved_in protein dephosphorylation 1 TAS Homo_sapiens(9606) Process GO:0008289 enables lipid binding 1 IEA Homo_sapiens(9606) Function GO:0010288 involved_in response to lead ion 1 ISS Homo_sapiens(9606) Process GO:0016791 enables phosphatase activity 1 IDA Homo_sapiens(9606) Function GO:0017018 enables myosin phosphatase activity 1 IEA Homo_sapiens(9606) Function GO:0032991 part_of protein-containing complex 1 IDA Homo_sapiens(9606) Component GO:0035970 involved_in peptidyl-threonine dephosphorylation 1 TAS Homo_sapiens(9606) Process GO:0042802 enables identical protein binding 1 IPI Homo_sapiens(9606) Function GO:0043123 involved_in positive regulation of I-kappaB kinase/NF-kappaB signaling 1 HMP Homo_sapiens(9606) Process GO:0043231 located_in intracellular membrane-bounded organelle 1 IDA Homo_sapiens(9606) Component GO:0043278 involved_in response to morphine 1 IEA Homo_sapiens(9606) Process GO:0043531 enables ADP binding 1 IDA Homo_sapiens(9606) Function GO:0046872 enables metal ion binding 1 IEA Homo_sapiens(9606) Function GO:0048156 enables tau protein binding 1 NAS Homo_sapiens(9606) Function GO:0051879 enables Hsp90 protein binding 1 IPI Homo_sapiens(9606) Function GO:0060090 enables molecular adaptor activity 1 EXP Homo_sapiens(9606) Function GO:0070262 involved_in peptidyl-serine dephosphorylation 2 IDA,TAS Homo_sapiens(9606) Process GO:0101031 part_of protein folding chaperone complex 1 IDA Homo_sapiens(9606) Component GO:1904550 involved_in response to arachidonic acid 1 ISS Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.