RBP Type
Non-canonical_RBPs
Diseases
Dengue
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
E3 ligase
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000005810 Gene ID
23077 Accession
23386
Symbol
MYCBP2
Alias
PAM;Phr;PHR1;Myc-bp2
Full Name
MYC binding protein 2
Status
Confidence
Length
284621 bases
Strand
Minus strand
Position
13 : 77042474 - 77327094
RNA binding domain
N.A.
Summary
This gene encodes an E3 ubiquitin-protein ligase and member of the PHR (Phr1/MYCBP2, highwire and RPM-1) family of proteins. The encoded protein plays a role in axon guidance and synapse formation in the developing nervous system. In mammalian cells, this protein regulates the cAMP and mTOR signaling pathways, and may additionally regulate autophagy. Reduced expression of this gene has been observed in acute lymphoblastic leukemia patients and a mutation in this gene has been identified in patients with a rare inherited vision defect. [provided by RefSeq, Mar 2017]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
MYCBP2-212
ENST00000695078
15315
No protein
-
MYCBP2-210
ENST00000683823
17326
4700aa
ENSP00000507196
MYCBP2-211
ENST00000684354
17128
4634aa
ENSP00000507330
MYCBP2-208
ENST00000682321
17482
4752aa
ENSP00000508023
MYCBP2-213
ENST00000695079
17221
4665aa
ENSP00000511683
MYCBP2-209
ENST00000683697
15359
4772aa
ENSP00000508153
MYCBP2-201
ENST00000357337
15257
4738aa
ENSP00000349892
MYCBP2-207
ENST00000544440
15077
4678aa
ENSP00000444596
MYCBP2-214
ENST00000695080
14539
4609aa
ENSP00000511684
MYCBP2-215
ENST00000695081
14323
4537aa
ENSP00000511685
MYCBP2-216
ENST00000695082
1861
No protein
-
MYCBP2-217
ENST00000695083
4922
No protein
-
MYCBP2-202
ENST00000429715
3660
1061aa
ENSP00000413907
MYCBP2-218
ENST00000695084
2855
No protein
-
MYCBP2-219
ENST00000695085
712
No protein
-
MYCBP2-220
ENST00000695086
8831
No protein
-
MYCBP2-221
ENST00000695087
1109
No protein
-
MYCBP2-205
ENST00000485061
566
No protein
-
MYCBP2-222
ENST00000695088
1729
No protein
-
MYCBP2-203
ENST00000466564
2402
No protein
-
MYCBP2-206
ENST00000486679
5088
No protein
-
MYCBP2-223
ENST00000695089
2885
No protein
-
MYCBP2-224
ENST00000695090
5985
No protein
-
MYCBP2-225
ENST00000695091
2638
No protein
-
MYCBP2-226
ENST00000695092
2106
No protein
-
MYCBP2-227
ENST00000695093
1019
No protein
-
MYCBP2-228
ENST00000695094
3181
No protein
-
MYCBP2-229
ENST00000695095
3214
No protein
-
MYCBP2-204
ENST00000474882
393
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
47528 Intelligence Tests 4.8295800E-005 - 47529 Intelligence Tests 4.8295800E-005 - 97720 Pre-Eclampsia 1E-6 23551011
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
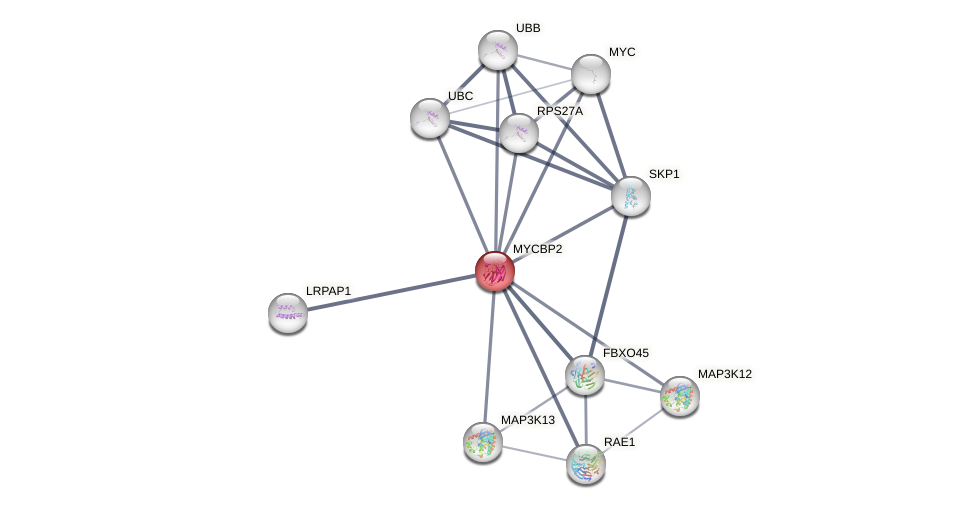
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0005085 enables guanyl-nucleotide exchange factor activity 1 IDA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 is_active_in nucleus 2 IBA,IDA Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 1 IDA Homo_sapiens(9606) Component GO:0005737 is_active_in cytoplasm 1 IC Homo_sapiens(9606) Component GO:0005829 located_in cytosol 1 IDA Homo_sapiens(9606) Component GO:0005886 is_active_in plasma membrane 1 IBA Homo_sapiens(9606) Component GO:0007411 involved_in axon guidance 1 IBA Homo_sapiens(9606) Process GO:0008270 enables zinc ion binding 1 IDA Homo_sapiens(9606) Function GO:0008582 involved_in regulation of synaptic assembly at neuromuscular junction 1 IBA Homo_sapiens(9606) Process GO:0015630 located_in microtubule cytoskeleton 1 IEA Homo_sapiens(9606) Component GO:0016020 located_in membrane 1 HDA Homo_sapiens(9606) Component GO:0016567 involved_in protein ubiquitination 2 IDA,IMP Homo_sapiens(9606) Process GO:0021785 involved_in branchiomotor neuron axon guidance 1 IEA Homo_sapiens(9606) Process GO:0021952 involved_in central nervous system projection neuron axonogenesis 1 ISS Homo_sapiens(9606) Process GO:0030424 located_in axon 1 IEA Homo_sapiens(9606) Component GO:0031267 enables small GTPase binding 1 IDA Homo_sapiens(9606) Function GO:0031398 involved_in positive regulation of protein ubiquitination 1 IDA Homo_sapiens(9606) Process GO:0032880 involved_in regulation of protein localization 1 IEA Homo_sapiens(9606) Process GO:0032922 involved_in circadian regulation of gene expression 1 IMP Homo_sapiens(9606) Process GO:0042177 involved_in negative regulation of protein catabolic process 1 IEA Homo_sapiens(9606) Process GO:0042802 enables identical protein binding 1 IEA Homo_sapiens(9606) Function GO:0043231 located_in intracellular membrane-bounded organelle 1 IDA Homo_sapiens(9606) Component GO:0050905 involved_in neuromuscular process 1 ISS Homo_sapiens(9606) Process GO:0051493 involved_in regulation of cytoskeleton organization 1 IEA Homo_sapiens(9606) Process GO:0061630 enables ubiquitin protein ligase activity 2 IBA,IDA Homo_sapiens(9606) Function GO:0070936 involved_in protein K48-linked ubiquitination 1 IDA Homo_sapiens(9606) Process GO:1902667 involved_in regulation of axon guidance 1 ISS Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.