RBP Type
Non-canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
E3 ligase
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000002746 Gene ID
23072 Accession
22195
Symbol
HECW1
Alias
NEDL1
Full Name
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
Status
Confidence
Length
453357 bases
Strand
Plus strand
Position
7 : 43112645 - 43566001
RNA binding domain
N.A.
Summary
Predicted to enable ubiquitin protein ligase activity. Predicted to be involved in several processes, including cellular protein metabolic process; negative regulation of sodium ion transmembrane transporter activity; and regulation of dendrite morphogenesis. Located in cytosol. [provided by Alliance of Genome Resources, Apr 2022]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
HECW1-201
ENST00000395891
9453
1606aa
ENSP00000379228
HECW1-209
ENST00000490954
531
No protein
-
HECW1-203
ENST00000453890
5169
1572aa
ENSP00000407774
HECW1-210
ENST00000492310
2186
No protein
-
HECW1-207
ENST00000471527
497
No protein
-
HECW1-211
ENST00000493057
750
No protein
-
HECW1-205
ENST00000464944
574
No protein
-
HECW1-208
ENST00000481031
412
No protein
-
HECW1-206
ENST00000471043
2888
No protein
-
HECW1-204
ENST00000461842
609
No protein
-
HECW1-202
ENST00000429529
415
138aa
ENSP00000413336
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
13251 Lupus Erythematosus, Systemic 1.17E-5 18204446 16835 Alcoholism 8.52E-05 - 16838 Alcoholism 6.14E-05 - 36729 Platelet Function Tests 1.5900000E-007 - 45254 Platelet Function Tests 8.0003842E-006 - 45295 Platelet Function Tests 3.5862709E-006 - 45300 Platelet Function Tests 3.4007331E-006 - 45380 Platelet Function Tests 1.8312700E-005 - 55860 lycopene 1.7740000E-005 - 55865 lycopene 2.8990000E-005 - 62361 Adenocarcinoma Of Esophagus 5E-6 24121790 67730 3-hydroxy-1-methylpropylmercapturic acid 1E-6 26053186 67731 Smoking 1E-6 26053186 74830 Bone Density 5E-6 27397699 79993 Alzheimer Disease 1E-6 23535033 90791 amyloid beta-protein (1-42) 3E-7 26252872 90792 Cerebrospinal Fluid 3E-7 26252872 102781 Immunoglobulin A 6E-6 28187132 104117 Energy Intake 6E-6 23251661 106473 Diabetes Mellitus, Type 2 4E-6 25102180
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
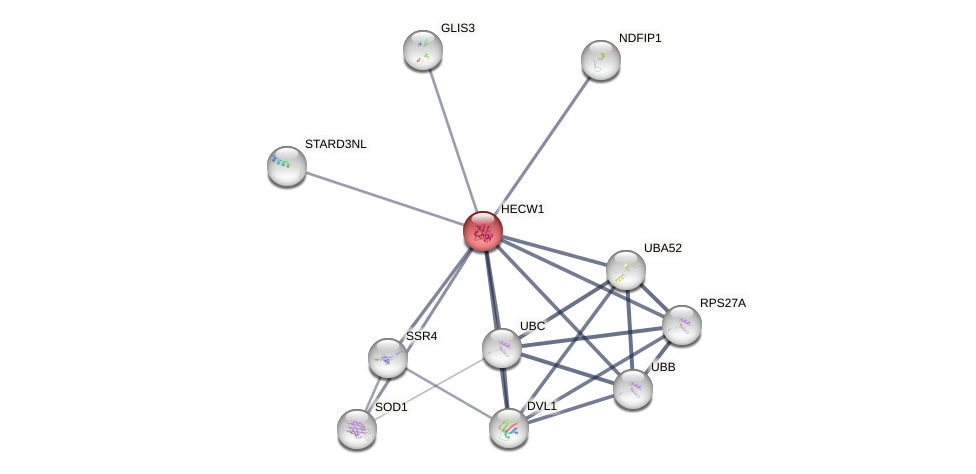
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005737 is_active_in cytoplasm 1 IBA Homo_sapiens(9606) Component GO:0005829 located_in cytosol 1 TAS Homo_sapiens(9606) Component GO:0016567 involved_in protein ubiquitination 2 IBA,IEA Homo_sapiens(9606) Process GO:0048814 involved_in regulation of dendrite morphogenesis 1 IBA Homo_sapiens(9606) Process GO:0061630 enables ubiquitin protein ligase activity 2 IBA,TAS Homo_sapiens(9606) Function GO:0090090 involved_in negative regulation of canonical Wnt signaling pathway 1 TAS Homo_sapiens(9606) Process GO:2000650 involved_in negative regulation of sodium ion transmembrane transporter activity 1 IBA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.