RBP Type
N.A.
Diseases
Drug
Main interacting RNAs
Moonlighting functions
Localizations
BulkPerturb-seq
Ensembl ID WBGene00006613 Gene ID
179920 Accession
-
Symbol
trm-1
Alias
NA
Full Name
tRNA (guanine(26)-N(2))-dimethyltransferase
Status
Confidence
Length
2490 bases
Strand
Minus strand
Position
V : 14183123 - 14185612
RNA binding domain
Met_10 , TRM
Summary
Predicted to enable tRNA (guanine-N2-)-methyltransferase activity. Predicted to be involved in tRNA N2-guanine methylation. Predicted to be active in nucleus. Human ortholog(s) of this gene implicated in autosomal recessive non-syndromic intellectual disability. Orthologous to human TRMT1 (tRNA methyltransferase 1). [provided by Alliance of Genome Resources, Apr 2022]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ZC376
Met_10
PF02475 9e-05
1
ZC376
Met_10
PF02475 9e-05
2
ZC376
TRM
PF02005 2.3e-154
3
ZC376
TRM
PF02005 2.3e-154
4
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
trm-1-201
ZC376
1827
526aa
ZC376
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
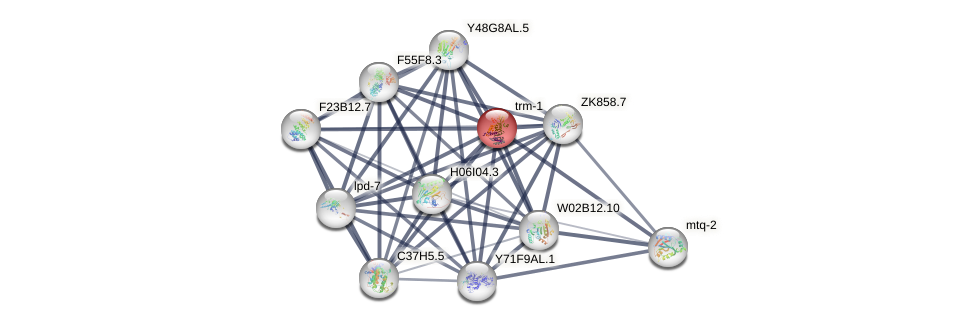
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
WBGene00006613
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
WBGene00006613 caenorhabditis_elegans ZC376.5.2 33.8403 18.0608 latimeria_chalumnae ENSLACP00000009578 26.1765 13.9706 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.9316 19.962 meleagris_gallopavo ENSMGAP00000028720 26.7705 14.8725 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.1711 19.962 cyclopterus_lumpus ENSCLMP00005017121 28.7714 16.3297 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.5513 19.3916 sphenodon_punctatus ENSSPUP00000005158 29.8246 16.2679 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.2205 18.4411 lepisosteus_oculatus ENSLOCP00000002063 25.5319 13.7589 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.6008 18.4411 petromyzon_marinus ENSPMAP00000006469 29.1667 15.5449 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.4525 20.1521 ornithorhynchus_anatinus ENSOANP00000001612 26.162 14.077 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.981 18.4411 laticauda_laticaudata ENSLLTP00000020797 25.3793 13.3793 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.5019 20.9125 anas_platyrhynchos_platyrhynchos ENSAPLP00000021906 21.0066 12.035 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.0304 19.0114 oryzias_latipes ENSORLP00000041756 26.8769 15.015 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.0304 18.0608 salmo_trutta ENSSTUP00000047723 25.7554 13.6691 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.6008 18.6312 danio_rerio ENSDARP00000114812 26.2248 14.121 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.6008 17.8707 esox_lucius ENSELUP00000021526 26.187 13.5252 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.3916 ailuropoda_melanoleuca ENSAMEP00000014299 26.0403 13.6913 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.1711 18.4411 ochotona_princeps ENSOPRP00000000620 28.3742 14.8773 WBGene00006613 caenorhabditis_elegans ZC376.5.2 33.0798 17.3004 ictalurus_punctatus ENSIPUP00000013032 28.5246 14.918 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.9316 19.962 gallus_gallus ENSGALP00010030573 25.3012 14.0562 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.4106 18.6312 haplochromis_burtoni ENSHBUP00000033518 26.8148 14.5185 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.4106 18.8213 astatotilapia_calliptera ENSACLP00000031962 26.8148 14.6667 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.692 19.5817 macaca_fascicularis ENSMFAP00000006895 26.3302 14.0518 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 macaca_nemestrina ENSMNEP00000037111 26.5306 14.0136 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.9316 20.3422 serinus_canaria ENSSCAP00000015109 24.4819 13.8601 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.6008 19.3916 poecilia_reticulata ENSPREP00000004059 26.8437 15.0442 WBGene00006613 caenorhabditis_elegans ZC376.5.2 31.9392 17.1103 echinops_telfairi ENSETEP00000012896 22.8571 12.2449 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.981 18.4411 pseudonaja_textilis ENSPTXP00000016019 25.1366 13.2514 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 papio_anubis ENSPANP00000010754 26.5306 14.0136 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.4106 19.0114 oreochromis_niloticus ENSONIP00000032826 24.7606 13.6799 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.1711 18.4411 dipodomys_ordii ENSDORP00000003083 28.4178 14.9002 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.7414 20.3422 parus_major ENSPMJP00000026271 25.5782 14.5578 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.2624 19.5817 vicugna_pacos ENSVPAP00000011031 30.0153 15.7734 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 mandrillus_leucophaeus ENSMLEP00000001222 26.5306 14.0136 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.4106 18.4411 clupea_harengus ENSCHAP00000037873 26.0057 13.9368 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.7719 panthera_leo ENSPLOP00000029035 26.8595 14.3251 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 loxodonta_africana ENSLAFP00000018950 26.4946 13.9946 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.7719 felis_catus ENSFCAP00000010483 26.8595 14.3251 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 cercocebus_atys ENSCATP00000041038 26.5306 14.0136 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.6426 19.7719 canis_lupus_familiaris ENSCAFP00845007120 26.4352 13.8852 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.5817 sus_scrofa ENSSSCP00000023028 23.9211 12.7004 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 bos_indicus_hybrid ENSBIXP00005024081 24.8724 13.1378 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.6426 19.7719 equus_asinus ENSEASP00005007804 23.8267 12.515 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.5019 20.7224 chrysemys_picta_bellii ENSCPBP00000021866 25.2632 14.3421 WBGene00006613 caenorhabditis_elegans ZC376.5.2 30.038 15.019 chinchilla_lanigera ENSCLAP00000015786 25.0794 12.5397 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.692 19.5817 chinchilla_lanigera ENSCLAP00000015714 26.7313 14.2659 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.981 19.0114 procavia_capensis ENSPCAP00000007735 25 13.587 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.6426 19.7719 vulpes_vulpes ENSVVUP00000012487 26.4352 13.8852 WBGene00006613 caenorhabditis_elegans ZC376.5.2 32.8897 17.6806 mus_spretus MGP_SPRETEiJ_P0021389 27.68 14.88 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.5019 19.5817 octodon_degus ENSODEP00000001035 27.2727 14.6307 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.6008 19.2015 sparus_aurata ENSSAUP00010016879 26.3386 14.6165 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.2205 18.4411 salmo_salar ENSSSAP00000153680 25.7143 13.8571 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.2624 20.3422 sarcophilus_harrisii ENSSHAP00000036158 25.5208 13.9323 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.3916 heterocephalus_glaber_female ENSHGLP00000004284 26.7857 14.011 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.6426 19.7719 equus_caballus ENSECAP00000018793 23.8267 12.515 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.2624 19.5817 cricetulus_griseus_chok1gshd ENSCGRP00001020025 26.8493 14.1096 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 pan_paniscus ENSPPAP00000011878 26.603 14.0518 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.0114 cavia_porcellus ENSCPOP00000001416 26.8698 13.8504 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.0304 18.0608 oncorhynchus_mykiss ENSOMYP00000072071 24.8957 13.2128 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.2205 19.2015 lates_calcarifer ENSLCAP00010019501 26.5487 14.8968 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.7909 18.8213 erinaceus_europaeus ENSEEUP00000012906 26.2931 14.2241 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 camelus_dromedarius ENSCDRP00005022007 26.603 14.0518 WBGene00006613 caenorhabditis_elegans ZC376.5.2 31.3688 16.73 sorex_araneus ENSSARP00000009174 30.3867 16.2063 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.7909 19.3916 crocodylus_porosus ENSCPRP00005018723 24.4 13.6 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.5817 physeter_catodon ENSPCTP00005004225 26.3587 13.9946 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 gorilla_gorilla ENSGGOP00000011489 26.603 14.0518 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 rhinopithecus_roxellana ENSRROP00000016053 26.5306 14.0136 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.4106 19.2015 xiphophorus_maculatus ENSXMAP00000003599 26.7751 14.9408 WBGene00006613 caenorhabditis_elegans ZC376.5.2 33.4601 18.0608 gadus_morhua ENSGMOP00000030585 25.5814 13.8081 WBGene00006613 caenorhabditis_elegans ZC376.5.2 32.6996 18.8213 oryzias_melastigma ENSOMEP00000014752 26.9592 15.5172 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 balaenoptera_musculus ENSBMSP00010001407 26.4946 13.9946 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.4106 19.5817 erpetoichthys_calabaricus ENSECRP00000019118 26.0807 14.8415 WBGene00006613 caenorhabditis_elegans ZC376.5.2 33.27 18.2509 dicentrarchus_labrax ENSDLAP00005013208 28.2258 15.4839 WBGene00006613 caenorhabditis_elegans ZC376.5.2 31.9392 16.9202 eptatretus_burgeri ENSEBUP00000016430 28.4264 15.0592 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.5817 urocitellus_parryii ENSUPAP00010004959 26.685 14.1678 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.7719 mus_pahari MGP_PahariEiJ_P0075612 26.7123 14.2466 WBGene00006613 caenorhabditis_elegans ZC376.5.2 27.1863 15.3992 neolamprologus_brichardi ENSNBRP00000005194 27.0321 15.3119 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 bos_grunniens ENSBGRP00000025255 24.8724 13.1378 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.692 19.7719 mus_spicilegus ENSMSIP00000010524 26.511 14.2857 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 delphinapterus_leucas ENSDLEP00000029086 24.9361 13.1714 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.3916 ursus_maritimus ENSUMAP00000030280 26.0403 13.6913 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.692 18.8213 dasypus_novemcinctus ENSDNOP00000004060 26.2943 13.4877 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 ovis_aries_rambouillet ENSOARP00020008112 26.387 13.9378 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.7414 19.7719 xenopus_tropicalis ENSXETP00000059823 27.2464 15.0725 WBGene00006613 caenorhabditis_elegans ZC376.5.2 28.8973 15.5894 tupaia_belangeri ENSTBEP00000014360 20.9366 11.2948 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.0304 19.7719 takifugu_rubripes ENSTRUP00000005540 25.035 14.5455 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.0304 18.0608 oncorhynchus_kisutch ENSOKIP00005092302 23.2166 12.3217 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.7719 panthera_pardus ENSPPRP00000001633 26.8595 14.3251 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.4106 18.8213 cyprinodon_variegatus ENSCVAP00000010118 26.6962 14.6018 WBGene00006613 caenorhabditis_elegans ZC376.5.2 23.1939 13.1179 choloepus_hoffmanni ENSCHOP00000009888 20.0328 11.33 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.5513 20.1521 ficedula_albicollis ENSFALP00000017486 25.6868 14.5604 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.2205 18.8213 pygocentrus_nattereri ENSPNAP00000021535 25.8993 14.2446 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.3916 tursiops_truncatus ENSTTRP00000005578 26.4305 13.8965 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.3916 myotis_lucifugus ENSMLUP00000001036 26.9819 14.1864 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.4525 19.3916 phocoena_sinus ENSPSNP00000005210 28.928 14.978 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 aotus_nancymaae ENSANAP00000041769 26.603 14.0518 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 bison_bison_bison ENSBBBP00000027144 26.749 14.1289 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.1217 20.3422 pelodiscus_sinensis ENSPSIP00000011148 27.1429 15.2857 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.6008 19.3916 oryzias_javanicus ENSOJAP00000001983 27.534 15.4312 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.7414 18.8213 mesocricetus_auratus ENSMAUP00000022466 26.2204 13.8075 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 rhinopithecus_bieti ENSRBIP00000038107 26.5306 14.0136 WBGene00006613 caenorhabditis_elegans ZC376.5.2 33.8403 18.8213 astyanax_mexicanus ENSAMXP00000009871 25.538 14.2037 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.7719 callithrix_jacchus ENSCJAP00000010090 26.8226 14.3054 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.1711 19.5817 kryptolebias_marmoratus ENSKMAP00000013762 28.5494 15.8951 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.962 rhinolophus_ferrumequinum ENSRFEP00010016965 26.7857 14.4231 WBGene00006613 caenorhabditis_elegans ZC376.5.2 33.6502 18.4411 electrophorus_electricus ENSEEEP00000010112 25.2137 13.8177 WBGene00006613 caenorhabditis_elegans ZC376.5.2 29.4677 16.73 tetraodon_nigroviridis ENSTNIP00000008233 36.9048 20.9524 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.1217 18.8213 anolis_carolinensis ENSACAP00000002310 26.3523 13.7309 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 capra_hircus ENSCHIP00000016225 26.387 13.9378 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.3612 18.6312 sinocyclocheilus_grahami ENSSGRP00000071082 28.7481 15.1468 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.2205 18.8213 sinocyclocheilus_grahami ENSSGRP00000083514 25.8993 14.2446 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.4106 18.8213 maylandia_zebra ENSMZEP00005023827 26.8148 14.6667 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 bos_mutus ENSBMUP00000029043 26.4586 13.9756 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.7719 panthera_tigris_altaica ENSPTIP00000026298 26.8595 14.3251 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.5817 saimiri_boliviensis_boliviensis ENSSBOP00000025626 26.4666 14.0518 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.5817 catagonus_wagneri ENSCWAP00000027132 25.4928 13.5348 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.4106 19.3916 seriola_dumerili ENSSDUP00000030264 26.9345 15.1786 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.2624 19.5817 cervus_hanglu_yarkandensis ENSCHYP00000013553 24.9682 13.121 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.9316 19.7719 phascolarctos_cinereus ENSPCIP00000002949 23.9544 13.1812 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 macaca_mulatta ENSMMUP00000018243 26.5306 14.0136 WBGene00006613 caenorhabditis_elegans ZC376.5.2 28.327 14.4487 propithecus_coquereli ENSPCOP00000001829 23.84 12.16 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.6008 19.3916 poecilia_formosa ENSPFOP00000004080 26.8437 15.0442 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.6008 19.5817 scophthalmus_maximus ENSSMAP00000025268 27.5758 15.6061 WBGene00006613 caenorhabditis_elegans ZC376.5.2 33.4601 17.8707 fundulus_heteroclitus ENSFHEP00000012863 32.9588 17.603 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.3612 18.2509 mustela_putorius_furo ENSMPUP00000011766 25.5494 13.1868 WBGene00006613 caenorhabditis_elegans ZC376.5.2 31.3688 16.73 carassius_auratus ENSCARP00000108219 25.076 13.3739 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.981 18.8213 carassius_auratus ENSCARP00000130738 26.6281 14.3271 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.981 18.6312 carassius_auratus ENSCARP00000122650 26.6281 14.1823 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.2205 18.6312 carassius_auratus ENSCARP00000082083 26.0116 14.1618 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 bos_taurus ENSBTAP00000063764 24.8724 13.1378 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 moschus_moschiferus ENSMMSP00000019123 24.8092 13.1043 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.0304 19.2015 anabas_testudineus ENSATEP00000006459 26.4012 14.8968 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.0304 18.2509 cynoglossus_semilaevis ENSCSEP00000001853 26.3623 14.1384 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 20.1521 vombatus_ursinus ENSVURP00010014773 27.6596 15.0355 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.2015 nannospalax_galili ENSNGAP00000008837 26.9071 14.0083 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.0114 jaculus_jaculus ENSJJAP00000006191 27.4399 14.1443 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.7414 18.6312 oryctolagus_cuniculus ENSOCUP00000004235 26.4045 13.764 WBGene00006613 caenorhabditis_elegans ZC376.5.2 31.7491 17.4905 amphilophus_citrinellus ENSACIP00000015529 34.0122 18.7373 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.7909 18.0608 scleropages_formosus ENSSFOP00015007392 26.4834 13.7482 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.2205 19.5817 amphiprion_ocellaris ENSAOCP00000028427 26.6272 15.2367 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.962 monodelphis_domestica ENSMODP00000009252 22.4277 12.1387 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.692 19.962 callorhinchus_milii ENSCMIP00000043753 26.0811 14.1892 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.9316 20.7224 gopherus_evgoodei ENSGEVP00005025256 24.9012 14.361 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.7719 mus_caroli MGP_CAROLIEiJ_P0020327 26.6484 14.2857 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.5817 ictidomys_tridecemlineatus ENSSTOP00000009280 26.685 14.1678 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 pongo_abelii ENSPPYP00000000472 26.603 14.0518 WBGene00006613 caenorhabditis_elegans ZC376.5.2 33.4601 19.2015 denticeps_clupeoides ENSDCDP00000016167 25.3237 14.5324 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.5019 20.3422 aquila_chrysaetos_chrysaetos ENSACCP00020009997 25.098 13.9869 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.5019 20.7224 strigops_habroptila ENSSHBP00005017136 25.5319 14.4947 WBGene00006613 caenorhabditis_elegans ZC376.5.2 33.4601 18.8213 betta_splendens ENSBSLP00000022430 26.6263 14.9773 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.1711 19.0114 gasterosteus_aculeatus ENSGACP00000006693 28.8162 15.5763 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.2205 19.3916 acanthochromis_polyacanthus ENSAPOP00000009427 26.6272 15.0888 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.6008 19.3916 poecilia_latipinna ENSPLAP00000024280 26.8437 15.0442 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.5019 19.2015 rattus_norvegicus ENSRNOP00000003517 27.3504 14.3875 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.2624 19.5817 peromyscus_maniculatus_bairdii ENSPEMP00000000030 27.1845 14.2857 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.5817 marmota_marmota_marmota ENSMMMP00000016480 26.685 14.1678 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.1217 20.7224 struthio_camelus_australis ENSSCUP00000006527 26.4256 15.1599 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 nomascus_leucogenys ENSNLEP00000021903 26.603 14.0518 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.3916 neovison_vison ENSNVIP00000012518 26.7857 14.011 WBGene00006613 caenorhabditis_elegans ZC376.5.2 30.038 15.2091 oncorhynchus_tshawytscha ENSOTSP00005052877 23.2695 11.782 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.7909 19.7719 sander_lucioperca ENSSLUP00000036617 26.5988 15.1163 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 monodon_monoceros ENSMMNP00015012999 26.4946 13.9946 WBGene00006613 caenorhabditis_elegans ZC376.5.2 57.4144 37.0722 saccharomyces_cerevisiae YDR120C 52.9825 34.2105 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.2624 19.2015 microcebus_murinus ENSMICP00000017297 26.7394 13.779 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.3612 19.962 geospiza_fortis ENSGFOP00000013483 26.8786 15.1734 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.7909 18.4411 notechis_scutatus ENSNSUP00000020112 28.6385 15.18 WBGene00006613 caenorhabditis_elegans ZC376.5.2 32.8897 18.2509 pundamilia_nyererei ENSPNYP00000007404 27.9483 15.5089 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.692 19.3916 ursus_americanus ENSUAMP00000027118 26.2585 13.8776 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 chlorocebus_sabaeus ENSCSAP00000009054 26.5306 14.0136 WBGene00006613 caenorhabditis_elegans ZC376.5.2 33.27 17.8707 hucho_hucho ENSHHUP00000074550 30.0687 16.1512 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 canis_lupus_dingo ENSCAFP00020019685 26.5306 14.0136 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.6426 19.962 microtus_ochrogaster ENSMOCP00000018057 27.3859 14.5228 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.7909 19.2015 leptobrachium_leishanense ENSLLEP00000048580 28.5491 15.7566 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.2205 18.8213 paramormyrops_kingsleyae ENSPKIP00000021612 25.825 14.2037 WBGene00006613 caenorhabditis_elegans ZC376.5.2 33.6502 18.6312 myripristis_murdjan ENSMMDP00005000436 28.32 15.68 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.7414 20.1521 coturnix_japonica ENSCJPP00005027796 25.1673 14.1901 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.981 18.6312 naja_naja ENSNNAP00000021801 25.1366 13.388 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.4106 19.2015 larimichthys_crocea ENSLCRP00005013399 26.7751 14.9408 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.0304 19.0114 oryzias_sinensis ENSOSIP00000024797 26.8769 15.015 WBGene00006613 caenorhabditis_elegans ZC376.5.2 29.8479 17.3004 labrus_bergylta ENSLBEP00000010299 25.6117 14.845 WBGene00006613 caenorhabditis_elegans ZC376.5.2 33.6502 18.8213 labrus_bergylta ENSLBEP00000004569 26.3001 14.7103 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.692 19.7719 cebus_imitator ENSCCAP00000003686 26.3302 14.1883 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.2205 19.0114 stegastes_partitus ENSSPAP00000011356 26.5879 14.771 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 homo_sapiens ENSP00000356476 26.603 14.0518 WBGene00006613 caenorhabditis_elegans ZC376.5.2 30.9886 16.73 podarcis_muralis ENSPMRP00000009293 36.3839 19.6429 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.0304 18.8213 mastacembelus_armatus ENSMAMP00000022858 24.7238 13.674 WBGene00006613 caenorhabditis_elegans ZC376.5.2 31.9392 17.6806 notamacropus_eugenii ENSMEUP00000010356 25.0373 13.8599 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.5817 sciurus_vulgaris ENSSVLP00005011333 26.7956 14.2265 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.2624 19.3916 carlito_syrichta ENSTSYP00000009593 27.1093 14.1079 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.5817 pan_troglodytes ENSPTRP00000002963 26.603 14.0518 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.3118 20.3422 taeniopygia_guttata ENSTGUP00000003867 24.9347 13.9687 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.9316 20.1521 chelonoidis_abingdonii ENSCABP00000003792 26.4336 14.8252 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.5019 20.7224 terrapene_carolina_triunguis ENSTMTP00000007472 25.2964 14.361 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.9316 20.3422 terrapene_carolina_triunguis ENSTMTP00000020361 28.4211 16.0902 WBGene00006613 caenorhabditis_elegans ZC376.5.2 33.8403 17.8707 pteropus_vampyrus ENSPVAP00000005635 24.3169 12.8415 WBGene00006613 caenorhabditis_elegans ZC376.5.2 37.0722 19.3916 prolemur_simus ENSPSMP00000019042 26.603 13.9154 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.8821 19.7719 mus_musculus ENSMUSP00000068309 26.6484 14.2857 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.9316 19.0114 salvator_merianae ENSSMRP00000016051 25.6445 13.5685 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.2205 19.5817 amphiprion_percula ENSAPEP00000030151 26.6272 15.2367 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.0304 18.8213 cyprinus_carpio_carpio ENSCCRP00000171361 25.7554 14.2446 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.7909 19.5817 cottoperca_gobio ENSCGOP00000000635 26.9514 15.1694 WBGene00006613 caenorhabditis_elegans ZC376.5.2 34.4106 19.3916 seriola_lalandi_dorsalis ENSSLDP00000021307 29.5752 16.6667 WBGene00006613 caenorhabditis_elegans ZC376.5.2 36.3118 20.5323 anser_brachyrhynchus ENSABRP00000003723 25.5007 14.4192 WBGene00006613 caenorhabditis_elegans ZC376.5.2 35.7414 19.0114 otolemur_garnettii ENSOGAP00000014497 30.1282 16.0256
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000049 enables tRNA binding 1 IEA Caenorhabditis_elegans(6239) Function GO:0002940 involved_in tRNA N2-guanine methylation 1 IBA Caenorhabditis_elegans(6239) Process GO:0003723 enables RNA binding 1 IEA Caenorhabditis_elegans(6239) Function GO:0004809 enables tRNA (guanine-N2-)-methyltransferase activity 2 IBA,IEA Caenorhabditis_elegans(6239) Function GO:0005634 is_active_in nucleus 1 IBA Caenorhabditis_elegans(6239) Component GO:0008033 involved_in tRNA processing 1 IEA Caenorhabditis_elegans(6239) Process GO:0008168 enables methyltransferase activity 1 IEA Caenorhabditis_elegans(6239) Function GO:0016740 enables transferase activity 1 IEA Caenorhabditis_elegans(6239) Function GO:0032259 involved_in methylation 1 IEA Caenorhabditis_elegans(6239) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.