Welcome to
RBP World!
Description
| Ensembl ID | WBGene00003014 | Gene ID | 172626 | Accession | - |
| Symbol | lin-28 | Alias | NA | Full Name | Protein lin-28 |
| Status | Confidence | Length | 3175 bases | Strand | Minus strand |
| Position | I : 8407630 - 8410804 | RNA binding domain | CSD , zf-CCHC | ||
| Summary | Enables RNA binding activity and enzyme binding activity. Involved in several processes, including regulation of cell fate specification; regulation of development, heterochronic; and regulation of gene expression. Located in cytoplasm and nucleus. Is expressed in several structures, including body wall musculature; nervous system; pharynx; reproductive system; and somatic gonad precursor. Orthologous to human LIN28A (lin-28 homolog A) and LIN28B (lin-28 homolog B). [provided by Alliance of Genome Resources, Apr 2022] | ||||
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| lin-28-201 | F02E9 | 1311 | 196aa | F02E9 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
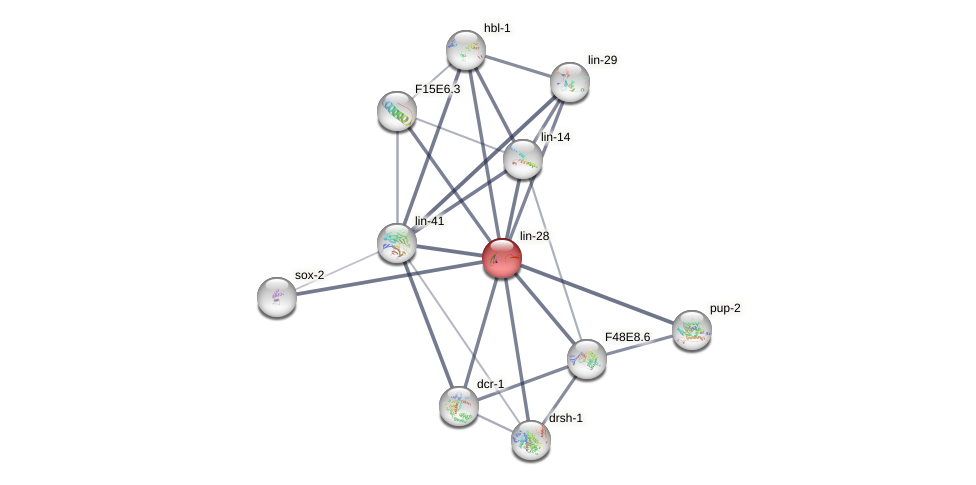
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| WBGene00003014 | ||||||||
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| WBGene00003014 | ||||||||
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0001708 | involved_in cell fate specification | 1 | IMP | Caenorhabditis_elegans(6239) | Process |
| GO:0003676 | enables nucleic acid binding | 1 | IEA | Caenorhabditis_elegans(6239) | Function |
| GO:0003729 | enables mRNA binding | 2 | IBA,IDA | Caenorhabditis_elegans(6239) | Function |
| GO:0003730 | enables mRNA 3-UTR binding | 1 | IDA | Caenorhabditis_elegans(6239) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Caenorhabditis_elegans(6239) | Function |
| GO:0005634 | is_active_in nucleus | 2 | IBA,IDA | Caenorhabditis_elegans(6239) | Component |
| GO:0005737 | is_active_in cytoplasm | 3 | IBA,IDA,IEA | Caenorhabditis_elegans(6239) | Component |
| GO:0007549 | involved_in dosage compensation | 1 | IGI | Caenorhabditis_elegans(6239) | Process |
| GO:0008270 | enables zinc ion binding | 1 | IEA | Caenorhabditis_elegans(6239) | Function |
| GO:0010629 | involved_in negative regulation of gene expression | 1 | IMP | Caenorhabditis_elegans(6239) | Process |
| GO:0019899 | enables enzyme binding | 2 | IEA,IPI | Caenorhabditis_elegans(6239) | Function |
| GO:0031054 | involved_in pre-miRNA processing | 2 | IBA,IDA | Caenorhabditis_elegans(6239) | Process |
| GO:0040034 | involved_in regulation of development, heterochronic | 2 | IGI,IMP | Caenorhabditis_elegans(6239) | Process |
| GO:0042659 | involved_in regulation of cell fate specification | 1 | IGI | Caenorhabditis_elegans(6239) | Process |
| GO:0046872 | enables metal ion binding | 1 | IEA | Caenorhabditis_elegans(6239) | Function |
| GO:0070878 | enables primary miRNA binding | 1 | IDA | Caenorhabditis_elegans(6239) | Function |
| GO:0070883 | enables pre-miRNA binding | 1 | IPI | Caenorhabditis_elegans(6239) | Function |
| GO:1990715 | enables mRNA CDS binding | 1 | IDA | Caenorhabditis_elegans(6239) | Function |
| GO:2000635 | involved_in negative regulation of primary miRNA processing | 1 | IMP | Caenorhabditis_elegans(6239) | Process |