| RBP Type |
N.A. |
| Diseases |
|
| Drug |
|
| Main interacting RNAs |
|
| Moonlighting functions |
|
| Localizations |
|
|
BulkPerturb-seq
|
|
| Ensembl ID | ENSNLEG00000001619 | Gene ID |
|
Accession |
- |
| Symbol |
ZC3H8 |
Alias |
- |
Full Name |
zinc finger CCCH-type containing 8 |
| Status |
Confidence |
Length |
40931 bases |
Strand |
Plus strand |
| Position |
14 : 75422425 - 75463355 |
RNA binding domain |
zf-CCCH , zf-CCCH_2 |
| Summary |
N.A. |
RNA binding domains (RBDs)

| Protein |
Domain |
Pfam ID |
E-value |
Domain number |
| ENSNLEP00000001922 |
zf-CCCH |
PF00642 |
1.1e-11 |
1 |
| ENSNLEP00000001922 |
zf-CCCH |
PF00642 |
1.1e-11 |
2 |
| ENSNLEP00000001922 |
zf-CCCH_2 |
PF14608 |
1.1e-09 |
3 |
| ENSNLEP00000001922 |
zf-CCCH_2 |
PF14608 |
1.1e-09 |
4 |
| ENSNLEP00000001922 |
zf-CCCH_2 |
PF14608 |
1.1e-09 |
5 |
RNA binding proteomes (RBPomes)
| Pubmed ID |
Full Name |
Cell |
Author |
Time |
Doi |
Literatures on RNA binding capacity
| Pubmed ID |
Title |
Author |
Time |
Journal |
| Name |
Transcript ID |
bp |
Protein |
Translation ID |
| ZC3H8-201 |
ENSNLET00000002032 |
1849 |
294aa |
ENSNLEP00000001922 |
| Pathway ID |
Pathway Name |
Source |
| | |
| ensgID |
Trait |
pValue |
Pubmed ID |
| ensgID | SNP | Chromosome | Position |
Trait | PubmedID | Or or BEAT |
EFO ID |
|---|
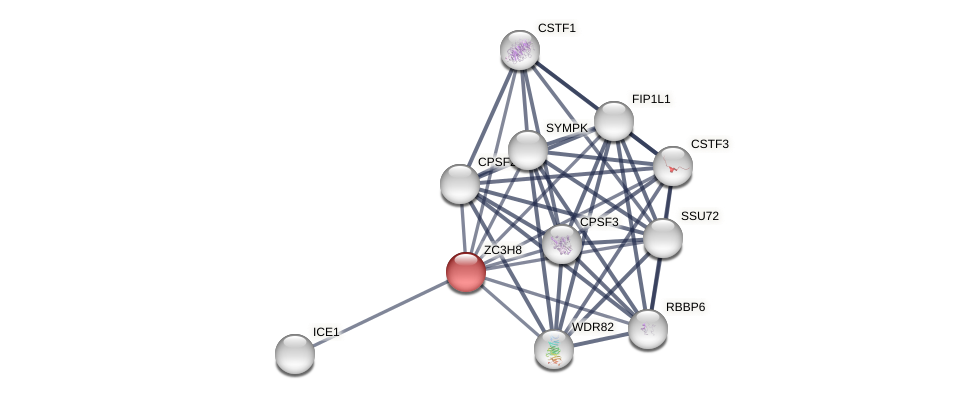
Protein-Protein Interaction (PPI)
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000037543 | 10.8306 | 6.67752 | nomascus_leucogenys | ENSNLEP00000001922 | 45.2381 | 27.8912 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000042659 | 15.2413 | 11.7697 | nomascus_leucogenys | ENSNLEP00000001922 | 61.2245 | 47.2789 |
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 97.619 | 96.9388 | pan_troglodytes | ENSPTRP00000021176 | 98.6254 | 97.9381 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 97.619 | 97.619 | homo_sapiens | ENSP00000386488 | 98.6254 | 98.6254 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 95.9184 | 94.5578 | pongo_abelii | ENSPPYP00000013563 | 93.3775 | 92.053 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 97.619 | 96.9388 | pan_paniscus | ENSPPAP00000009677 | 98.6254 | 97.9381 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 97.2789 | 96.5986 | gorilla_gorilla | ENSGGOP00000008803 | 98.2818 | 97.5945 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 69.3878 | 65.3061 | choloepus_hoffmanni | ENSCHOP00000009201 | 74.1818 | 69.8182 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 83.6735 | 77.2109 | erinaceus_europaeus | ENSEEUP00000006352 | 82.5503 | 76.1745 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 67.0068 | 60.2041 | echinops_telfairi | ENSETEP00000013437 | 66.5541 | 59.7973 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 86.7347 | 84.3537 | callithrix_jacchus | ENSCJAP00000078371 | 86.4407 | 84.0678 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 94.5578 | 93.8775 | cercocebus_atys | ENSCATP00000014534 | 93.2886 | 92.6174 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 96.9388 | 96.2585 | macaca_fascicularis | ENSMFAP00000016722 | 96.6102 | 95.9322 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 96.5986 | 96.2585 | macaca_mulatta | ENSMMUP00000041412 | 96.2712 | 95.9322 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 96.9388 | 96.2585 | macaca_nemestrina | ENSMNEP00000021502 | 96.6102 | 95.9322 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 96.5986 | 95.9184 | papio_anubis | ENSPANP00000009595 | 96.2712 | 95.5932 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 96.2585 | 95.5782 | mandrillus_leucophaeus | ENSMLEP00000011615 | 95.9322 | 95.2542 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 90.4762 | 87.415 | canis_lupus_familiaris | ENSCAFP00845014428 | 80.8511 | 78.1155 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 90.1361 | 87.415 | vulpes_vulpes | ENSVVUP00000040130 | 86.8852 | 84.2623 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 86.7347 | 82.9932 | ursus_americanus | ENSUAMP00000002656 | 83.0619 | 79.4788 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 87.415 | 83.6735 | ailuropoda_melanoleuca | ENSAMEP00000026000 | 83.1715 | 79.6116 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 91.8367 | 89.1156 | felis_catus | ENSFCAP00000007496 | 86.8167 | 84.2444 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 88.4354 | 85.3742 | mustela_putorius_furo | ENSMPUP00000009485 | 83.6013 | 80.7074 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 91.8367 | 89.1156 | panthera_leo | ENSPLOP00000000371 | 86.8167 | 84.2444 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 87.0748 | 84.6939 | panthera_pardus | ENSPPRP00000011936 | 83.3876 | 81.1075 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 78.9116 | 75.5102 | tursiops_truncatus | ENSTTRP00000012480 | 75.3247 | 72.0779 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 86.7347 | 81.9728 | delphinapterus_leucas | ENSDLEP00000006409 | 80.4416 | 76.0252 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 58.5034 | 56.1224 | physeter_catodon | ENSPCTP00005011549 | 86.8687 | 83.3333 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 85.7143 | 81.2925 | balaenoptera_musculus | ENSBMSP00010026145 | 88.4211 | 83.8596 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 88.7755 | 83.6735 | loxodonta_africana | ENSLAFP00000003902 | 86.1386 | 81.1881 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 92.8571 | 89.7959 | equus_caballus | ENSECAP00000020190 | 89.2157 | 86.2745 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 92.517 | 89.4558 | equus_asinus | ENSEASP00005041984 | 88.8889 | 85.9477 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 66.3265 | 60.2041 | procavia_capensis | ENSPCAP00000014399 | 64.5695 | 58.6093 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 90.1361 | 86.7347 | camelus_dromedarius | ENSCDRP00005013835 | 86.6013 | 83.3333 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 87.7551 | 82.3129 | sus_scrofa | ENSSSCP00000057275 | 83.4951 | 78.3172 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 88.0952 | 79.932 | bos_taurus | ENSBTAP00000021230 | 85.1974 | 77.3026 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 86.0544 | 79.2517 | capra_hircus | ENSCHIP00000009201 | 84.0532 | 77.4086 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 86.3946 | 80.2721 | ovis_aries_rambouillet | ENSOARP00020002484 | 80.6349 | 74.9206 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 77.8912 | 71.0884 | ochotona_princeps | ENSOPRP00000010207 | 82.3741 | 75.1799 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 85.7143 | 80.2721 | oryctolagus_cuniculus | ENSOCUP00000028419 | 70.9859 | 66.4789 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 85.3742 | 80.9524 | marmota_marmota_marmota | ENSMMMP00000002478 | 89.6429 | 85 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 93.1973 | 88.0952 | urocitellus_parryii | ENSUPAP00010011915 | 89.5425 | 84.6405 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 88.7755 | 81.2925 | dipodomys_ordii | ENSDORP00000021355 | 85.8553 | 78.6184 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 86.7347 | 79.2517 | cricetulus_griseus_chok1gshd | ENSCGRP00001018685 | 83.3333 | 76.1438 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 85.3742 | 78.2313 | mesocricetus_auratus | ENSMAUP00000012106 | 82.0261 | 75.1634 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 87.7551 | 79.5918 | mus_caroli | MGP_CAROLIEiJ_P0054518 | 84.5902 | 76.7213 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 88.4354 | 80.6122 | mus_musculus | ENSMUSP00000028866 | 85.2459 | 77.7049 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 87.7551 | 79.5918 | mus_pahari | MGP_PahariEiJ_P0065496 | 84.5902 | 76.7213 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 87.7551 | 79.932 | mus_spretus | MGP_SPRETEiJ_P0056626 | 84.5902 | 77.0492 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 82.9932 | 76.1905 | rattus_norvegicus | ENSRNOP00000023845 | 80 | 73.4426 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 80.2721 | 72.449 | cavia_porcellus | ENSCPOP00000007424 | 84.2857 | 76.0714 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 81.2925 | 73.1292 | mus_spicilegus | ENSMSIP00000021247 | 81.017 | 72.8814 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 89.7959 | 80.9524 | heterocephalus_glaber_female | ENSHGLP00000011673 | 86.2745 | 77.7778 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 89.7959 | 86.0544 | ursus_maritimus | ENSUMAP00000011526 | 86.5574 | 82.9508 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 87.0748 | 77.551 | octodon_degus | ENSODEP00000008825 | 83.6601 | 74.5098 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 87.7551 | 79.932 | bos_grunniens | ENSBGRP00000042775 | 84.8684 | 77.3026 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 88.0952 | 79.932 | bos_indicus_hybrid | ENSBIXP00005012989 | 85.4785 | 77.5578 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 90.1361 | 86.7347 | vicugna_pacos | ENSVPAP00000005487 | 87.1711 | 83.8816 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 94.898 | 92.1769 | microcebus_murinus | ENSMICP00000048845 | 91.1765 | 88.5621 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 88.7755 | 80.9524 | chinchilla_lanigera | ENSCLAP00000009888 | 85.5738 | 78.0328 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 90.4762 | 86.7347 | otolemur_garnettii | ENSOGAP00000002284 | 85.5305 | 81.9936 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 50 | 48.6395 | tupaia_belangeri | ENSTBEP00000013490 | 55.4717 | 53.9623 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 80.6122 | 78.2313 | aotus_nancymaae | ENSANAP00000008214 | 80.6122 | 78.2313 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 88.4354 | 86.0544 | saimiri_boliviensis_boliviensis | ENSSBOP00000023321 | 84.9673 | 82.6797 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 90.4762 | 86.3946 | monodon_monoceros | ENSMMNP00015027589 | 86.0841 | 82.2006 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 91.1565 | 87.0748 | phocoena_sinus | ENSPSNP00000020938 | 79.5252 | 75.9644 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 93.5374 | 88.4354 | ictidomys_tridecemlineatus | ENSSTOP00000005672 | 89.8693 | 84.9673 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 88.4354 | 80.2721 | bison_bison_bison | ENSBBBP00000001565 | 85.5263 | 77.6316 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 90.8163 | 86.3946 | catagonus_wagneri | ENSCWAP00000013879 | 86.129 | 81.9355 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 76.8708 | 69.3878 | sorex_araneus | ENSSARP00000011647 | 80.427 | 72.5979 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 88.0952 | 79.2517 | jaculus_jaculus | ENSJJAP00000001725 | 84.918 | 76.3934 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 78.2313 | 71.7687 | myotis_lucifugus | ENSMLUP00000004453 | 83.0325 | 76.1733 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 92.517 | 87.0748 | sciurus_vulgaris | ENSSVLP00005008667 | 89.1803 | 83.9344 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 83.3333 | 82.6531 | chlorocebus_sabaeus | ENSCSAP00000012854 | 96.0784 | 95.2941 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 86.3946 | 80.6122 | rhinolophus_ferrumequinum | ENSRFEP00010021811 | 78.1538 | 72.9231 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 96.5986 | 96.2585 | rhinopithecus_roxellana | ENSRROP00000031531 | 96.2712 | 95.9322 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 87.0748 | 84.6939 | panthera_tigris_altaica | ENSPTIP00000018660 | 83.3876 | 81.1075 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 96.5986 | 95.9184 | rhinopithecus_bieti | ENSRBIP00000007040 | 96.2712 | 95.5932 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 88.4354 | 85.3742 | canis_lupus_dingo | ENSCAFP00020022190 | 78.5499 | 75.8308 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 85.034 | 77.551 | microtus_ochrogaster | ENSMOCP00000016018 | 82.2368 | 75 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 82.3129 | 74.4898 | nannospalax_galili | ENSNGAP00000007110 | 75.8621 | 68.652 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 67.0068 | 63.9456 | pteropus_vampyrus | ENSPVAP00000003320 | 66.5541 | 63.5135 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 85.7143 | 78.9116 | peromyscus_maniculatus_bairdii | ENSPEMP00000009921 | 83.4437 | 76.8212 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 82.9932 | 80.6122 | neovison_vison | ENSNVIP00000021825 | 82.9932 | 80.6122 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 95.2381 | 91.8367 | propithecus_coquereli | ENSPCOP00000019483 | 91.5033 | 88.2353 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 89.7959 | 88.7755 | cebus_imitator | ENSCCAP00000004527 | 89.7959 | 88.7755 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 88.7755 | 87.7551 | cebus_imitator | ENSCCAP00000032510 | 92.5532 | 91.4894 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 87.415 | 78.2313 | cervus_hanglu_yarkandensis | ENSCHYP00000013623 | 82.9032 | 74.1936 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 89.1156 | 79.932 | cervus_hanglu_yarkandensis | ENSCHYP00000018897 | 86.1842 | 77.3026 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 87.0748 | 82.6531 | prolemur_simus | ENSPSMP00000023451 | 79.7508 | 75.7009 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 89.1156 | 80.6122 | moschus_moschiferus | ENSMMSP00000021429 | 86.1842 | 77.9605 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 93.5374 | 89.4558 | carlito_syrichta | ENSTSYP00000006110 | 90.4605 | 86.5132 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 88.0952 | 80.2721 | bos_mutus | ENSBMUP00000032730 | 85.1974 | 77.6316 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 32.9932 | 20.4082 | caenorhabditis_elegans | T26A8.4.2 | 16.899 | 10.453 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 38.4354 | 23.4694 | ciona_intestinalis | ENSCINP00000034981 | 11.566 | 7.06244 |
| ENSNLEG00000001619 | nomascus_leucogenys | ENSNLEP00000001922 | 36.7347 | 23.1293 | hippocampus_comes | ENSHCOP00000017324 | 12.3147 | 7.75371 |
| Go ID |
Go term |
No. evidence |
Entries |
Species |
Category |