| RBP Type |
N.A. |
| Diseases |
|
| Drug |
|
| Main interacting RNAs |
|
| Moonlighting functions |
|
| Localizations |
|
|
BulkPerturb-seq
|
|
| Ensembl ID | ENSNLEG00000001420 | Gene ID |
|
Accession |
- |
| Symbol |
CPSF4L |
Alias |
- |
Full Name |
cleavage and polyadenylation specific factor 4 like |
| Status |
Confidence |
Length |
13488 bases |
Strand |
Plus strand |
| Position |
14 : 51600667 - 51614154 |
RNA binding domain |
zf-CCCH , zf-CCCH_2 |
| Summary |
N.A. |
RNA binding domains (RBDs)

| Protein |
Domain |
Pfam ID |
E-value |
Domain number |
| ENSNLEP00000001680 |
zf-CCCH |
PF00642 |
6.4e-18 |
1 |
| ENSNLEP00000001680 |
zf-CCCH_2 |
PF14608 |
2.2e-09 |
2 |
| ENSNLEP00000001680 |
zf-CCCH_2 |
PF14608 |
2.2e-09 |
3 |
| ENSNLEP00000001680 |
zf-CCCH_2 |
PF14608 |
2.2e-09 |
4 |
| ENSNLEP00000001680 |
zf-CCCH_2 |
PF14608 |
2.2e-09 |
5 |
| ENSNLEP00000001680 |
zf-CCCH_2 |
PF14608 |
2.2e-09 |
6 |
| ENSNLEP00000028571 |
zf-CCCH |
PF00642 |
5.8e-12 |
1 |
| ENSNLEP00000028571 |
zf-CCCH_2 |
PF14608 |
7.4e-10 |
2 |
| ENSNLEP00000028571 |
zf-CCCH_2 |
PF14608 |
7.4e-10 |
3 |
| ENSNLEP00000028571 |
zf-CCCH_2 |
PF14608 |
7.4e-10 |
4 |
| ENSNLEP00000028571 |
zf-CCCH_2 |
PF14608 |
7.4e-10 |
5 |
RNA binding proteomes (RBPomes)
| Pubmed ID |
Full Name |
Cell |
Author |
Time |
Doi |
Literatures on RNA binding capacity
| Pubmed ID |
Title |
Author |
Time |
Journal |
| Name |
Transcript ID |
bp |
Protein |
Translation ID |
| CPSF4L-201 |
ENSNLET00000001778 |
643 |
179aa |
ENSNLEP00000001680 |
| CPSF4L-202 |
ENSNLET00000038974 |
459 |
152aa |
ENSNLEP00000028571 |
| Pathway ID |
Pathway Name |
Source |
| | |
| ensgID |
Trait |
pValue |
Pubmed ID |
| ensgID | SNP | Chromosome | Position |
Trait | PubmedID | Or or BEAT |
EFO ID |
|---|
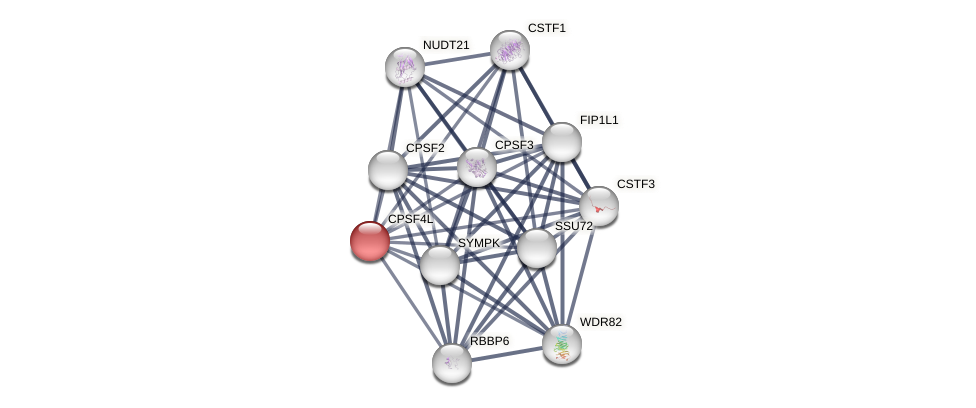
Protein-Protein Interaction (PPI)
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001996 | 8.5197 | 5.43131 | nomascus_leucogenys | ENSNLEP00000001680 | 44.6927 | 28.4916 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000029704 | 51.7241 | 44.0613 | nomascus_leucogenys | ENSNLEP00000001680 | 75.419 | 64.2458 |
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 99.4413 | 99.4413 | pan_troglodytes | ENSPTRP00000016303 | 99.4413 | 99.4413 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 99.4413 | 99.4413 | homo_sapiens | ENSP00000343900 | 99.4413 | 99.4413 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 98.8827 | 98.324 | pongo_abelii | ENSPPYP00000009644 | 98.8827 | 98.324 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 55.8659 | 55.8659 | pan_paniscus | ENSPPAP00000027868 | 65.7895 | 65.7895 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 99.4413 | 99.4413 | gorilla_gorilla | ENSGGOP00000028251 | 99.4413 | 99.4413 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 77.095 | 69.8324 | sarcophilus_harrisii | ENSSHAP00000026532 | 79.7688 | 72.2543 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 49.162 | 43.5754 | notamacropus_eugenii | ENSMEUP00000003682 | 86.2745 | 76.4706 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 88.2682 | 83.2402 | dasypus_novemcinctus | ENSDNOP00000011202 | 82.2917 | 77.6042 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 98.324 | 96.0894 | cercocebus_atys | ENSCATP00000044115 | 98.324 | 96.0894 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 98.324 | 96.0894 | macaca_mulatta | ENSMMUP00000001695 | 98.324 | 96.0894 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 98.324 | 96.0894 | macaca_nemestrina | ENSMNEP00000045883 | 98.324 | 96.0894 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 83.7989 | 80.4469 | papio_anubis | ENSPANP00000037754 | 66.0793 | 63.4361 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 98.8827 | 96.648 | mandrillus_leucophaeus | ENSMLEP00000028599 | 98.8827 | 96.648 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 53.0726 | 47.486 | canis_lupus_familiaris | ENSCAFP00845027181 | 53.9773 | 48.2955 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 67.5978 | 62.5698 | vulpes_vulpes | ENSVVUP00000013205 | 74.6914 | 69.1358 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 74.3017 | 68.1564 | ursus_americanus | ENSUAMP00000009271 | 84.7134 | 77.707 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 87.1508 | 79.3296 | ailuropoda_melanoleuca | ENSAMEP00000002852 | 93.9759 | 85.5422 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 33.5196 | 30.7263 | felis_catus | ENSFCAP00000058025 | 65.9341 | 60.4396 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 62.5698 | 56.9832 | felis_catus | ENSFCAP00000004386 | 51.8519 | 47.2222 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 83.7989 | 75.9777 | mustela_putorius_furo | ENSMPUP00000014169 | 89.2857 | 80.9524 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 55.8659 | 51.9553 | panthera_leo | ENSPLOP00000008280 | 41.841 | 38.9121 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 67.0391 | 60.3352 | panthera_pardus | ENSPPRP00000005213 | 67.4157 | 60.6742 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 54.1899 | 51.3966 | tursiops_truncatus | ENSTTRP00000004940 | 64.2384 | 60.9272 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 58.1006 | 54.1899 | delphinapterus_leucas | ENSDLEP00000016517 | 47.7064 | 44.4954 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 87.7095 | 83.2402 | physeter_catodon | ENSPCTP00005010997 | 94.5783 | 89.759 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 90.5028 | 85.4749 | balaenoptera_musculus | ENSBMSP00010023277 | 89.011 | 84.0659 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 84.9162 | 77.6536 | loxodonta_africana | ENSLAFP00000029284 | 86.3636 | 78.9773 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 89.3855 | 84.3575 | equus_caballus | ENSECAP00000089371 | 72.0721 | 68.018 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 89.3855 | 84.3575 | equus_asinus | ENSEASP00005036169 | 72.0721 | 68.018 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 88.2682 | 83.7989 | camelus_dromedarius | ENSCDRP00005020033 | 95.1807 | 90.3614 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 62.5698 | 54.7486 | sus_scrofa | ENSSSCP00000057357 | 51.3761 | 44.9541 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 61.4525 | 55.3073 | capra_hircus | ENSCHIP00000025069 | 71.8954 | 64.7059 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 52.514 | 45.2514 | ovis_aries_rambouillet | ENSOARP00020033877 | 62.2517 | 53.6424 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 53.0726 | 48.0447 | ochotona_princeps | ENSOPRP00000006847 | 70.3704 | 63.7037 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 82.6816 | 75.9777 | oryctolagus_cuniculus | ENSOCUP00000018443 | 85.5491 | 78.6127 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 88.8268 | 80.4469 | marmota_marmota_marmota | ENSMMMP00000011463 | 71.6216 | 64.8649 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 86.5922 | 79.3296 | urocitellus_parryii | ENSUPAP00010003139 | 78.2828 | 71.7172 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 83.2402 | 73.743 | cricetulus_griseus_chok1gshd | ENSCGRP00001021159 | 67.7273 | 60 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 82.6816 | 73.743 | mesocricetus_auratus | ENSMAUP00000021942 | 89.1566 | 79.5181 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 82.6816 | 70.9497 | mus_caroli | MGP_CAROLIEiJ_P0030777 | 64.9123 | 55.7018 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 82.6816 | 70.9497 | mus_musculus | ENSMUSP00000097819 | 52.8571 | 45.3571 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 39.6648 | 28.4916 | mus_pahari | MGP_PahariEiJ_P0037504 | 24.9123 | 17.8947 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 83.2402 | 72.067 | mus_spretus | MGP_SPRETEiJ_P0032226 | 65.3509 | 56.5789 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 56.9832 | 48.6034 | rattus_norvegicus | ENSRNOP00000074846 | 66.6667 | 56.8627 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 55.3073 | 46.3687 | mus_spicilegus | ENSMSIP00000012204 | 52.381 | 43.9153 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 74.3017 | 68.1564 | ursus_maritimus | ENSUMAP00000010458 | 84.7134 | 77.707 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 49.7207 | 43.0168 | monodelphis_domestica | ENSMODP00000049827 | 51.1494 | 44.2529 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 78.2123 | 70.9497 | monodelphis_domestica | ENSMODP00000038500 | 84.3373 | 76.506 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 89.3855 | 85.4749 | microcebus_murinus | ENSMICP00000006832 | 91.954 | 87.931 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 75.419 | 69.8324 | vombatus_ursinus | ENSVURP00010016083 | 81.8182 | 75.7576 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 43.5754 | 40.7821 | tupaia_belangeri | ENSTBEP00000010999 | 86.6667 | 81.1111 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 75.419 | 70.9497 | phascolarctos_cinereus | ENSPCIP00000002336 | 81.8182 | 76.9697 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 36.8715 | 35.7542 | aotus_nancymaae | ENSANAP00000017357 | 49.6241 | 48.1203 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 67.0391 | 60.8939 | saimiri_boliviensis_boliviensis | ENSSBOP00000035565 | 75.9494 | 68.9873 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 58.1006 | 54.7486 | monodon_monoceros | ENSMMNP00015020843 | 47.7064 | 44.9541 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 88.8268 | 85.4749 | phocoena_sinus | ENSPSNP00000030097 | 95.7831 | 92.1687 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 87.1508 | 80.4469 | ictidomys_tridecemlineatus | ENSSTOP00000001648 | 89.6552 | 82.7586 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 84.9162 | 76.5363 | myotis_lucifugus | ENSMLUP00000008750 | 84.9162 | 76.5363 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 55.8659 | 48.0447 | sciurus_vulgaris | ENSSVLP00005011632 | 61.3497 | 52.7607 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 97.7654 | 95.5307 | chlorocebus_sabaeus | ENSCSAP00000000764 | 97.2222 | 95 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 79.8883 | 75.9777 | rhinolophus_ferrumequinum | ENSRFEP00010012523 | 87.7301 | 83.4356 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 97.2067 | 94.9721 | rhinopithecus_roxellana | ENSRROP00000026609 | 97.2067 | 94.9721 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 77.095 | 71.5084 | panthera_tigris_altaica | ENSPTIP00000012692 | 84.6626 | 78.5276 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 92.7374 | 88.2682 | rhinopithecus_bieti | ENSRBIP00000010391 | 91.2088 | 86.8132 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 54.1899 | 46.9274 | canis_lupus_dingo | ENSCAFP00020027420 | 45.5399 | 39.4366 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 69.2737 | 60.3352 | microtus_ochrogaster | ENSMOCP00000016401 | 85.5172 | 74.4828 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 84.3575 | 76.5363 | nannospalax_galili | ENSNGAP00000006291 | 88.3041 | 80.117 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 51.9553 | 48.0447 | pteropus_vampyrus | ENSPVAP00000006359 | 62.4161 | 57.7181 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 75.419 | 64.8045 | peromyscus_maniculatus_bairdii | ENSPEMP00000035009 | 67.1642 | 57.7114 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 86.5922 | 79.8883 | neovison_vison | ENSNVIP00000011838 | 93.3735 | 86.1446 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 89.3855 | 84.9162 | propithecus_coquereli | ENSPCOP00000019685 | 90.9091 | 86.3636 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 60.3352 | 55.3073 | cebus_imitator | ENSCCAP00000025581 | 80 | 73.3333 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 94.4134 | 89.3855 | prolemur_simus | ENSPSMP00000037273 | 94.4134 | 89.3855 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 65.3631 | 53.6313 | carlito_syrichta | ENSTSYP00000017447 | 74.0506 | 60.7595 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 64.8045 | 50.2793 | caenorhabditis_elegans | F11A10.8.1 | 38.4106 | 29.8013 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 43.5754 | 34.6369 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000020789 | 45.8824 | 36.4706 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 5.58659 | 3.91061 | meleagris_gallopavo | ENSMGAP00000013727 | 1.86916 | 1.30841 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 45.8101 | 29.6089 | gallus_gallus | ENSGALP00010011567 | 8.143 | 5.26316 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 59.7765 | 50.2793 | serinus_canaria | ENSSCAP00000018498 | 33.9683 | 28.5714 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 6.14525 | 3.35196 | pelodiscus_sinensis | ENSPSIP00000009072 | 2.00364 | 1.0929 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 53.6313 | 37.4302 | saccharomyces_cerevisiae | YPR107C | 46.1538 | 32.2115 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 46.3687 | 30.1676 | coturnix_japonica | ENSCJPP00005012571 | 8.76452 | 5.70222 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 73.1844 | 62.5698 | aquila_chrysaetos_chrysaetos | ENSACCP00020010201 | 79.8781 | 68.2927 |
| ENSNLEG00000001420 | nomascus_leucogenys | ENSNLEP00000001680 | 71.5084 | 59.2179 | strigops_habroptila | ENSSHBP00005016971 | 79.0123 | 65.4321 |
| Go ID |
Go term |
No. evidence |
Entries |
Species |
Category |