RBP Type
N.A.
Diseases
Drug
Main interacting RNAs
Moonlighting functions
Localizations
BulkPerturb-seq
Ensembl ID ENSMUSG00000058594 Gene ID
50755 Accession
-
Symbol
Fbh1
Alias
Fbx18;Fbxo18
Full Name
F-box DNA helicase 1
Status
Confidence
Length
35008 bases
Strand
Minus strand
Position
2 : 11747386 - 11782393
RNA binding domain
UvrD-helicase
Summary
Predicted to enable ATP-dependent activity, acting on DNA and DNA binding activity. Involved in negative regulation of double-strand break repair via homologous recombination; replication fork processing; and replication fork protection. Acts upstream of or within negative regulation of chromatin binding activity and response to intra-S DNA damage checkpoint signaling. Predicted to be located in chromatin and nucleus. Predicted to be part of SCF ubiquitin ligase complex. Orthologous to human FBH1 (F-box DNA helicase 1). [provided by Alliance of Genome Resources, Apr 2022]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSMUSP00000071495
UvrD-helicase
PF00580 2e-11
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
Fbh1-207
ENSMUST00000139292
358
No protein
-
Fbh1-201
ENSMUST00000071564
3567
1042aa
ENSMUSP00000071495
Fbh1-209
ENSMUST00000155604
1064
No protein
-
Fbh1-202
ENSMUST00000123717
4118
No protein
-
Fbh1-205
ENSMUST00000131893
891
297aa
ENSMUSP00000116392
Fbh1-206
ENSMUST00000136633
374
No protein
-
Fbh1-203
ENSMUST00000123723
820
No protein
-
Fbh1-210
ENSMUST00000192171
476
No protein
-
Fbh1-204
ENSMUST00000126543
898
No protein
-
Fbh1-208
ENSMUST00000151402
503
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
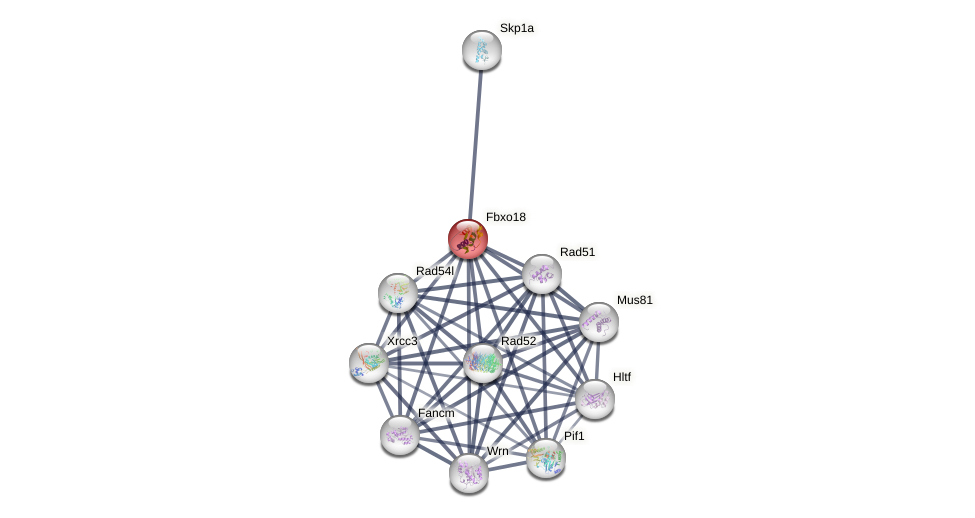
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSMUSG00000058594
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSMUSG00000058594
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000166 enables nucleotide binding 1 IEA Mus_musculus(10090) Function GO:0000724 involved_in double-strand break repair via homologous recombination 1 IBA Mus_musculus(10090) Process GO:0000785 part_of chromatin 1 ISO Mus_musculus(10090) Component GO:0001934 acts_upstream_of_or_within positive regulation of protein phosphorylation 1 ISO Mus_musculus(10090) Process GO:0003677 enables DNA binding 1 IEA Mus_musculus(10090) Function GO:0003678 enables DNA helicase activity 1 ISO Mus_musculus(10090) Function GO:0003690 enables double-stranded DNA binding 1 ISO Mus_musculus(10090) Function GO:0003697 enables single-stranded DNA binding 1 ISO Mus_musculus(10090) Function GO:0004386 enables helicase activity 1 IEA Mus_musculus(10090) Function GO:0005515 enables protein binding 1 IPI Mus_musculus(10090) Function GO:0005524 enables ATP binding 1 IEA Mus_musculus(10090) Function GO:0005634 is_active_in nucleus 2 IBA,ISO Mus_musculus(10090) Component GO:0005694 located_in chromosome 1 IEA Mus_musculus(10090) Component GO:0006281 involved_in DNA repair 1 ISO Mus_musculus(10090) Process GO:0006308 acts_upstream_of_or_within DNA catabolic process 1 ISO Mus_musculus(10090) Process GO:0006974 involved_in DNA damage response 1 ISO Mus_musculus(10090) Process GO:0015616 enables DNA translocase activity 1 ISO Mus_musculus(10090) Function GO:0016567 involved_in protein ubiquitination 1 ISO Mus_musculus(10090) Process GO:0016787 enables hydrolase activity 1 IEA Mus_musculus(10090) Function GO:0016853 enables isomerase activity 1 IEA Mus_musculus(10090) Function GO:0019005 part_of SCF ubiquitin ligase complex 1 ISO Mus_musculus(10090) Component GO:0031297 involved_in replication fork processing 3 IBA,IMP,ISO Mus_musculus(10090) Process GO:0035562 acts_upstream_of_or_within negative regulation of chromatin binding 1 IMP Mus_musculus(10090) Process GO:0043138 enables 3-5 DNA helicase activity 2 IBA,ISO Mus_musculus(10090) Function GO:0072429 acts_upstream_of_or_within response to intra-S DNA damage checkpoint signaling 2 IMP,ISO Mus_musculus(10090) Process GO:1902231 acts_upstream_of_or_within positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage 1 ISO Mus_musculus(10090) Process GO:2000042 involved_in negative regulation of double-strand break repair via homologous recombination 2 IMP,ISO Mus_musculus(10090) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.