RBP Type
N.A.
Diseases
Drug
Main interacting RNAs
Moonlighting functions
Localizations
BulkPerturb-seq
Ensembl ID ENSMUSG00000037331 Gene ID
73158 Accession
-
Symbol
Larp1
Alias
Larp;mKIAA0731;1810024J12Rik;3110040D16Rik
Full Name
La ribonucleoprotein 1, translational regulator
Status
Confidence
Length
52971 bases
Strand
Plus strand
Position
11 : 57899890 - 57952860
RNA binding domain
La
Summary
Predicted to enable eukaryotic initiation factor 4E binding activity; nucleic acid binding activity; and ribosomal small subunit binding activity. Predicted to be involved in several processes, including TORC1 signaling; cellular response to rapamycin; and posttranscriptional regulation of gene expression. Predicted to act upstream of or within regulation of translation. Predicted to be located in cytoplasm. Predicted to be part of TORC1 complex and polysome. Predicted to be active in cytoplasmic stress granule and cytosol. Predicted to colocalize with polysomal ribosome. Is expressed in several structures, including branchial arch; cranial ganglion; epithelium; future spinal cord; and limb bud. Orthologous to human LARP1 (La ribonucleoprotein 1, translational regulator). [provided by Alliance of Genome Resources, Apr 2022]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSMUSP00000071421
La
PF05383 4.3e-20
1
ENSMUSP00000136673
La
PF05383 4.5e-20
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
Larp1-201
ENSMUST00000071487
6614
1072aa
ENSMUSP00000071421
Larp1-202
ENSMUST00000140500
747
No protein
-
Larp1-203
ENSMUST00000178636
6617
1072aa
ENSMUSP00000136673
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
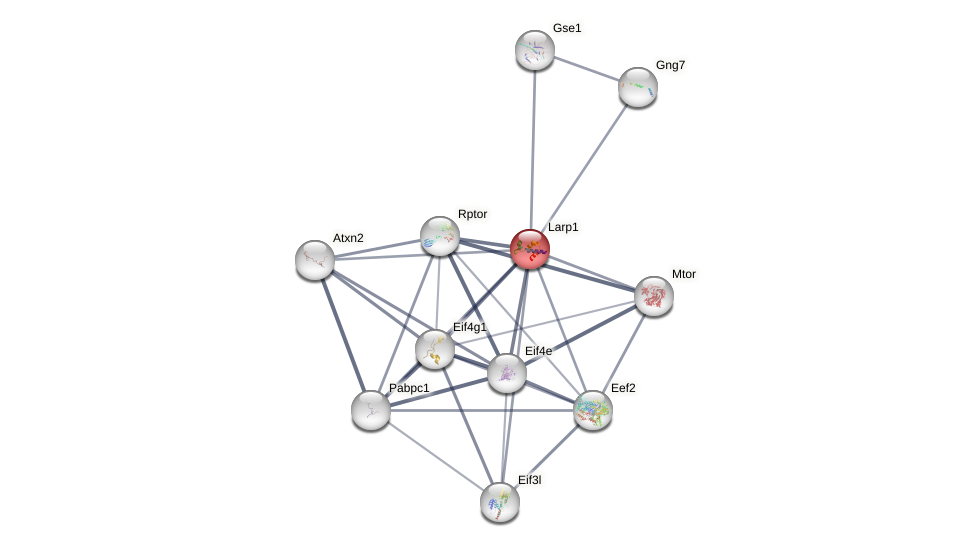
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSMUSG00000037331 [{"sID": "ENSMUSG00000037331" "sSP": "mus_musculus" "sProtein": "ENSMUSP00000136673" "sPercPos": 12.4067 "sPercID": 6.43657 "tID": "ENSMUSG00000068882" "tSP": "mus_musculus" "tGene": "Ssb"
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSMUSG00000037331
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000339 enables RNA cap binding 1 ISO Mus_musculus(10090) Function GO:0000340 enables RNA 7-methylguanosine cap binding 1 ISO Mus_musculus(10090) Function GO:0003723 enables RNA binding 1 IBA Mus_musculus(10090) Function GO:0003730 enables mRNA 3-UTR binding 1 ISO Mus_musculus(10090) Function GO:0005737 located_in cytoplasm 2 ISA,ISO Mus_musculus(10090) Component GO:0005829 is_active_in cytosol 1 IBA Mus_musculus(10090) Component GO:0005844 part_of polysome 1 ISO Mus_musculus(10090) Component GO:0006413 involved_in translational initiation 1 ISO Mus_musculus(10090) Process GO:0006417 acts_upstream_of_or_within regulation of translation 1 IEA Mus_musculus(10090) Process GO:0008190 enables eukaryotic initiation factor 4E binding 1 ISO Mus_musculus(10090) Function GO:0008283 involved_in cell population proliferation 1 ISO Mus_musculus(10090) Process GO:0008494 enables translation activator activity 1 ISO Mus_musculus(10090) Function GO:0010494 is_active_in cytoplasmic stress granule 2 IBA,ISO Mus_musculus(10090) Component GO:0010608 involved_in post-transcriptional regulation of gene expression 1 ISO Mus_musculus(10090) Process GO:0016239 involved_in positive regulation of macroautophagy 1 ISO Mus_musculus(10090) Process GO:0017148 involved_in negative regulation of translation 1 ISO Mus_musculus(10090) Process GO:0031369 enables translation initiation factor binding 1 ISO Mus_musculus(10090) Function GO:0031929 involved_in TOR signaling 1 ISO Mus_musculus(10090) Process GO:0031931 part_of TORC1 complex 1 ISO Mus_musculus(10090) Component GO:0038202 involved_in TORC1 signaling 1 ISO Mus_musculus(10090) Process GO:0042788 colocalizes_with polysomal ribosome 1 ISO Mus_musculus(10090) Component GO:0043024 enables ribosomal small subunit binding 1 ISO Mus_musculus(10090) Function GO:0045070 involved_in positive regulation of viral genome replication 1 ISO Mus_musculus(10090) Process GO:0045727 involved_in positive regulation of translation 1 IBA Mus_musculus(10090) Process GO:0045947 involved_in negative regulation of translational initiation 1 ISO Mus_musculus(10090) Process GO:0045948 involved_in positive regulation of translational initiation 1 ISO Mus_musculus(10090) Process GO:0048027 enables mRNA 5-UTR binding 1 ISO Mus_musculus(10090) Function GO:0048255 involved_in mRNA stabilization 1 ISO Mus_musculus(10090) Process GO:0072752 involved_in cellular response to rapamycin 1 ISO Mus_musculus(10090) Process GO:1990928 involved_in response to amino acid starvation 1 ISO Mus_musculus(10090) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.