RBP Type
N.A.
Diseases
Drug
Main interacting RNAs
Moonlighting functions
Localizations
BulkPerturb-seq
Ensembl ID ENSMUSG00000032560 Gene ID
235567 Accession
-
Symbol
Dnajc13
Alias
Rme8;RME-8;Gm1124;mKIAA0678;D030002L11Rik
Full Name
DnaJ heat shock protein family (Hsp40) member C13
Status
Confidence
Length
111649 bases
Strand
Minus strand
Position
9 : 104028481 - 104140129
RNA binding domain
GYF_2
Summary
Predicted to be involved in endosome organization; receptor-mediated endocytosis; and regulation of cytoplasmic transport. Predicted to be located in cytosol and early endosome membrane. Predicted to be part of WASH complex. Predicted to be active in endosome membrane. Is expressed in thymus primordium. Human ortholog(s) of this gene implicated in Parkinson's disease. Orthologous to human DNAJC13 (DnaJ heat shock protein family (Hsp40) member C13). [provided by Alliance of Genome Resources, Apr 2022]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSMUSP00000140571
GYF_2
PF14237 2.1e-17
1
ENSMUSP00000035170
GYF_2
PF14237 4.4e-16
1
ENSMUSP00000139804
GYF_2
PF14237 4.4e-16
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
Dnajc13-201
ENSMUST00000035170
8012
2243aa
ENSMUSP00000035170
Dnajc13-211
ENSMUST00000189813
601
No protein
-
Dnajc13-204
ENSMUST00000188139
668
96aa
ENSMUSP00000150468
Dnajc13-213
ENSMUST00000190926
453
No protein
-
Dnajc13-203
ENSMUST00000186788
6747
2248aa
ENSMUSP00000139804
Dnajc13-207
ENSMUST00000188885
492
59aa
ENSMUSP00000141056
Dnajc13-208
ENSMUST00000188998
614
184aa
ENSMUSP00000140867
Dnajc13-205
ENSMUST00000188425
3237
No protein
-
Dnajc13-214
ENSMUST00000191199
615
No protein
-
Dnajc13-202
ENSMUST00000185503
1222
174aa
ENSMUSP00000140571
Dnajc13-209
ENSMUST00000189103
641
No protein
-
Dnajc13-206
ENSMUST00000188592
734
No protein
-
Dnajc13-212
ENSMUST00000190600
526
No protein
-
Dnajc13-210
ENSMUST00000189299
568
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
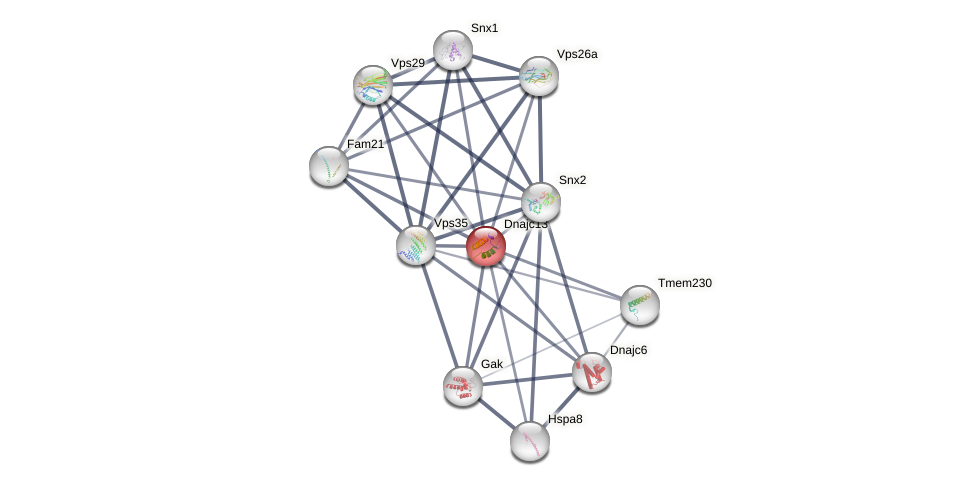
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSMUSG00000032560 [{"sID": "ENSMUSG00000032560" "sSP": "mus_musculus" "sProtein": "ENSMUSP00000139804" "sPercPos": 6.58363 "sPercID": 3.6032 "tID": "ENSMUSG00000014195" "tSP": "mus_musculus" "tGene": "NA"
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSMUSG00000032560
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0003674 enables molecular_function 1 ND Mus_musculus(10090) Function GO:0005829 located_in cytosol 1 ISO Mus_musculus(10090) Component GO:0006898 involved_in receptor-mediated endocytosis 1 IBA Mus_musculus(10090) Process GO:0007032 involved_in endosome organization 1 ISO Mus_musculus(10090) Process GO:0010008 is_active_in endosome membrane 2 IBA,ISO Mus_musculus(10090) Component GO:0031901 located_in early endosome membrane 1 ISO Mus_musculus(10090) Component GO:0043231 located_in intracellular membrane-bounded organelle 1 ISO Mus_musculus(10090) Component GO:0071203 part_of WASH complex 1 ISO Mus_musculus(10090) Component GO:1902954 involved_in regulation of early endosome to recycling endosome transport 1 ISO Mus_musculus(10090) Process GO:2000641 involved_in regulation of early endosome to late endosome transport 1 ISO Mus_musculus(10090) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.