RBP Type
N.A.
Diseases
Drug
Main interacting RNAs
Moonlighting functions
Localizations
BulkPerturb-seq
Ensembl ID ENSMUSG00000024782 Gene ID
56248 Accession
-
Symbol
Ak3
Alias
AK-3;Ak3l;Ak3l1;Akl3l
Full Name
adenylate kinase 3
Status
Confidence
Length
27129 bases
Strand
Minus strand
Position
19 : 28998233 - 29025361
RNA binding domain
ADK_lid
Summary
Predicted to enable adenylate kinase activity; identical protein binding activity; and nucleoside triphosphate adenylate kinase activity. Predicted to be involved in ADP biosynthetic process and nucleoside triphosphate metabolic process. Predicted to act upstream of or within phosphorylation. Located in mitochondrial matrix. Is expressed in several structures, including alimentary system; genitourinary system; nervous system; respiratory system; and sensory organ. Orthologous to human AK3 (adenylate kinase 3). [provided by Alliance of Genome Resources, Apr 2022]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSMUSP00000121867
ADK_lid
PF05191 4.1e-17
1
ENSMUSP00000025696
ADK_lid
PF05191 3.9e-16
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
Ak3-201
ENSMUST00000025696
2809
227aa
ENSMUSP00000025696
Ak3-202
ENSMUST00000125587
504
65aa
ENSMUSP00000121867
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
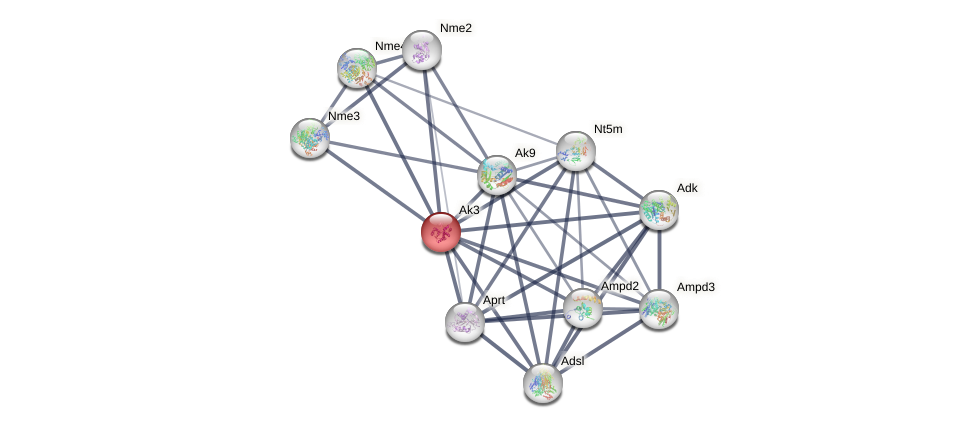
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSMUSG00000024782
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSMUSG00000024782
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000166 enables nucleotide binding 1 IEA Mus_musculus(10090) Function GO:0004017 enables adenylate kinase activity 2 IBA,ISO Mus_musculus(10090) Function GO:0004550 enables nucleoside diphosphate kinase activity 1 IEA Mus_musculus(10090) Function GO:0005524 enables ATP binding 1 IEA Mus_musculus(10090) Function GO:0005525 enables GTP binding 1 IEA Mus_musculus(10090) Function GO:0005737 is_active_in cytoplasm 1 IBA Mus_musculus(10090) Component GO:0005739 located_in mitochondrion 3 HDA,IDA,ISO Mus_musculus(10090) Component GO:0005759 is_active_in mitochondrial matrix 3 IBA,IDA,ISO Mus_musculus(10090) Component GO:0006139 involved_in nucleobase-containing compound metabolic process 1 IEA Mus_musculus(10090) Process GO:0006172 involved_in ADP biosynthetic process 1 ISO Mus_musculus(10090) Process GO:0006756 involved_in AMP phosphorylation 1 ISO Mus_musculus(10090) Process GO:0009142 involved_in nucleoside triphosphate biosynthetic process 1 IBA Mus_musculus(10090) Process GO:0016301 enables kinase activity 1 IEA Mus_musculus(10090) Function GO:0016310 acts_upstream_of_or_within phosphorylation 1 IEA Mus_musculus(10090) Process GO:0016740 enables transferase activity 1 IEA Mus_musculus(10090) Function GO:0016776 enables phosphotransferase activity, phosphate group as acceptor 1 IEA Mus_musculus(10090) Function GO:0019205 enables nucleobase-containing compound kinase activity 1 IEA Mus_musculus(10090) Function GO:0042802 enables identical protein binding 1 ISO Mus_musculus(10090) Function GO:0046033 involved_in AMP metabolic process 2 IBA,ISO Mus_musculus(10090) Process GO:0046039 involved_in GTP metabolic process 1 ISO Mus_musculus(10090) Process GO:0046041 involved_in ITP metabolic process 1 ISO Mus_musculus(10090) Process GO:0046051 involved_in UTP metabolic process 1 ISO Mus_musculus(10090) Process GO:0046060 involved_in dATP metabolic process 1 ISO Mus_musculus(10090) Process GO:0046899 enables nucleoside triphosphate adenylate kinase activity 2 IBA,ISO Mus_musculus(10090) Function
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.