Welcome to
RBP World!
Functions and Diseases
| RBP Type | Non-canonical_RBPs |
| Diseases | Cancer Disease |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | TF |
| Localizations | Stress granucleP-body |
| BulkPerturb-seq | DataSet_02_21 |
Description
| Ensembl ID | ENSG00000214575 | Gene ID | 64506 | Accession | 21744 |
| Symbol | CPEB1 | Alias | CPEB;CPEB-1;h-CPEB;CPE-BP1;hCPEB-1 | Full Name | cytoplasmic polyadenylation element binding protein 1 |
| Status | Confidence | Length | 104777 bases | Strand | Minus strand |
| Position | 15 : 82543201 - 82647977 | RNA binding domain | N.A. | ||
| Summary | This gene encodes a member of the cytoplasmic polyadenylation element binding protein family. This highly conserved protein binds to a specific RNA sequence, called the cytoplasmic polyadenylation element, found in the 3' untranslated region of some mRNAs. The encoded protein functions in both the cytoplasm and the nucleus. It is involved in the regulation of mRNA translation, as well as processing of the 3' untranslated region, and may play a role in cell proliferation and tumorigenesis. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2014] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 27495319 | Gld2-catalyzed 3' monoadenylation of miRNAs in the hippocampus has no detectable effect on their stability or on animal behavior. | Fernanda Mansur | 2016-10-01 | RNA (New York, N.Y.) |
| 35217597 | Oppositional poly(A) tail length regulation by FMRP and CPEB1. | Jihae Shin | 2022-05-01 | RNA (New York, N.Y.) |
| 25216517 | CPEB1 modulates differentiation of glioma stem cells via downregulation of HES1 and SIRT1 expression. | Jinlong Yin | 2014-08-30 | Oncotarget |
| 26493705 | Non-coding functions of alternative pre-mRNA splicing in development. | Stefan Mockenhaupt | 2015-12-01 | Seminars in cell & developmental biology |
| 30237545 | CPEB1 mediates hepatocellular carcinoma cancer stemness and chemoresistance. | Min Xu | 2018-09-20 | Cell death & disease |
| 35177647 | CPEB1 directs muscle stem cell activation by reprogramming the translational landscape. | Wenshu Zeng | 2022-02-17 | Nature communications |
| 32968053 | CPEB3-mediated MTDH mRNA translational suppression restrains hepatocellular carcinoma progression. | He Zhang | 2020-09-23 | Cell death & disease |
| 28798901 | Translational Dysregulation in Cancer: Molecular Insights and Potential Clinical Applications in Biomarker Development. | Christos Vaklavas | 2017-01-01 | Frontiers in oncology |
| 33228708 | miR-129-3p alleviates chondrocyte apoptosis in knee joint fracture-induced osteoarthritis through CPEB1. | Ruixiong Chen | 2020-11-23 | Journal of orthopaedic surgery and research |
| 36510219 | Re-evaluation of the myoepithelial cells roles in the breast cancer progression. | Anwar Shams | 2022-12-12 | Cancer cell international |
| 37340420 | Virus usurps alternative splicing to clear the decks for infection. | Ruixue Li | 2023-06-20 | Virology journal |
| 27271671 | A Multi-Step miRNA-mRNA Regulatory Network Construction Approach Identifies Gene Signatures Associated with Endometrioid Endometrial Carcinoma. | Hanzhen Xiong | 2016-06-02 | Genes |
| 36768153 | Autism Spectrum Disorder: Neurodevelopmental Risk Factors, Biological Mechanism, and Precision Therapy. | Ling Wang | 2023-01-17 | International journal of molecular sciences |
| 23942453 | Restarting life: fertilization and the transition from meiosis to mitosis. | Dean Clift | 2013-09-01 | Nature reviews. Molecular cell biology |
| 24990967 | A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins. | Tariq Afroz | 2014-07-01 | Genes & development |
| 31034770 | Viral modulation of cellular RNA alternative splicing: A new key player in virus-host interactions? | Simon Boudreault | 2019-09-01 | Wiley interdisciplinary reviews. RNA |
| 36092352 | Low CPEB1 levels may predict the benefit of 5-fluorouracil treatment in patients with colon or stomach adenocarcinoma. | Jing-Zhu Cao | 2022-08-01 | Journal of gastrointestinal oncology |
| 28383716 | Essential role for non-canonical poly(A) polymerase GLD4 in cytoplasmic polyadenylation and carbohydrate metabolism. | Jihae Shin | 2017-06-20 | Nucleic acids research |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| CPEB1-217 | ENST00000618698 | 2885 | No protein | - |
| CPEB1-209 | ENST00000614918 | 4882 | 588aa | ENSP00000481010 |
| CPEB1-216 | ENST00000618449 | 2459 | 491aa | ENSP00000483590 |
| CPEB1-215 | ENST00000617958 | 2776 | 626aa | ENSP00000478598 |
| CPEB1-211 | ENST00000615198 | 2112 | 561aa | ENSP00000477715 |
| CPEB1-208 | ENST00000611163 | 1919 | 486aa | ENSP00000483857 |
| CPEB1-212 | ENST00000616959 | 2009 | 564aa | ENSP00000479625 |
| CPEB1-213 | ENST00000617462 | 2068 | 486aa | ENSP00000477557 |
| CPEB1-218 | ENST00000620182 | 2422 | 486aa | ENSP00000484549 |
| CPEB1-214 | ENST00000617522 | 2220 | 486aa | ENSP00000481009 |
| CPEB1-210 | ENST00000614977 | 1559 | 138aa | ENSP00000478841 |
| CPEB1-206 | ENST00000570205 | 569 | No protein | - |
| CPEB1-204 | ENST00000568994 | 554 | 114aa | ENSP00000455044 |
| CPEB1-205 | ENST00000569257 | 527 | 35aa | ENSP00000455268 |
| CPEB1-203 | ENST00000567678 | 557 | 17aa | ENSP00000457137 |
| CPEB1-202 | ENST00000566716 | 502 | No protein | - |
| CPEB1-201 | ENST00000563519 | 536 | No protein | - |
| CPEB1-207 | ENST00000611031 | 3074 | 561aa | ENSP00000483951 |
| CPEB1-219 | ENST00000684509 | 3482 | 588aa | ENSP00000507835 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
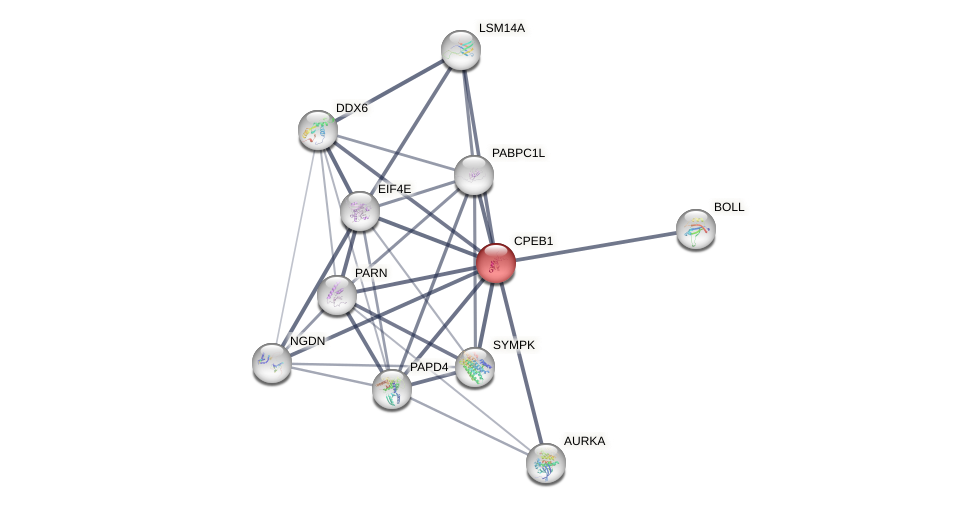
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000214575 | homo_sapiens | ENSP00000443985 | 24.4681 | 16.441 | homo_sapiens | ENSP00000507835 | 43.0272 | 28.9116 |
| ENSG00000214575 | homo_sapiens | ENSP00000265997 | 35.3868 | 23.4957 | homo_sapiens | ENSP00000507835 | 42.0068 | 27.8912 |
| ENSG00000214575 | homo_sapiens | ENSP00000265085 | 36.6255 | 24.2798 | homo_sapiens | ENSP00000507835 | 45.4082 | 30.102 |
| ENSG00000214575 | homo_sapiens | ENSP00000488102 | 91.0543 | 90.0958 | homo_sapiens | ENSP00000507835 | 96.9388 | 95.9184 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.7483 | 94.2177 | nomascus_leucogenys | ENSNLEP00000036709 | 91.5447 | 90.0813 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.2381 | 94.0476 | pan_paniscus | ENSPPAP00000001719 | 89.4569 | 88.3387 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.5782 | 94.2177 | pongo_abelii | ENSPPYP00000029873 | 90.0641 | 88.7821 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.4082 | 94.2177 | pan_troglodytes | ENSPTRP00000081191 | 89.6166 | 88.4984 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 79.4218 | 78.0612 | gorilla_gorilla | ENSGGOP00000037707 | 88.1132 | 86.6038 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 85.8844 | 80.6122 | ornithorhynchus_anatinus | ENSOANP00000021085 | 95.6439 | 89.7727 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 85.2041 | 82.3129 | sarcophilus_harrisii | ENSSHAP00000016124 | 95.977 | 92.7203 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 64.966 | 61.7347 | notamacropus_eugenii | ENSMEUP00000015099 | 82.684 | 78.5714 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 44.0476 | 43.0272 | choloepus_hoffmanni | ENSCHOP00000004142 | 56.1822 | 54.8807 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 88.6054 | 88.0952 | dasypus_novemcinctus | ENSDNOP00000017129 | 99.6176 | 99.044 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.3878 | 93.7075 | callithrix_jacchus | ENSCJAP00000029634 | 98.2301 | 97.5221 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 96.5986 | 94.5578 | cercocebus_atys | ENSCATP00000008893 | 91.6129 | 89.6774 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 79.4218 | 77.8912 | macaca_mulatta | ENSMMUP00000058357 | 79.9658 | 78.4247 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.7483 | 94.2177 | macaca_nemestrina | ENSMNEP00000023354 | 91.8434 | 90.3752 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 99.8299 | 98.9796 | papio_anubis | ENSPANP00000059591 | 99.8299 | 98.9796 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 96.5986 | 94.5578 | mandrillus_leucophaeus | ENSMLEP00000038958 | 91.6129 | 89.6774 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.2177 | 92.8571 | tursiops_truncatus | ENSTTRP00000012346 | 98.7522 | 97.3262 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 93.8775 | 91.1565 | vulpes_vulpes | ENSVVUP00000007876 | 91.6944 | 89.0365 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 90.6463 | 89.4558 | mus_spretus | MGP_SPRETEiJ_P0083814 | 94.8399 | 93.5943 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.7279 | 93.7075 | equus_asinus | ENSEASP00005009785 | 98.4099 | 97.3498 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.2177 | 93.0272 | capra_hircus | ENSCHIP00000031781 | 97.7072 | 96.4727 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 92.3469 | 90.6463 | loxodonta_africana | ENSLAFP00000016062 | 97.3118 | 95.5197 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.898 | 93.7075 | panthera_leo | ENSPLOP00000022384 | 99.4652 | 98.2175 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 90.3061 | 87.9252 | ailuropoda_melanoleuca | ENSAMEP00000024679 | 78.3186 | 76.2537 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.2381 | 93.5374 | bos_indicus_hybrid | ENSBIXP00005035931 | 92.869 | 91.2106 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 87.7551 | 84.8639 | oryctolagus_cuniculus | ENSOCUP00000028690 | 79.0199 | 76.4165 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.5782 | 94.2177 | equus_caballus | ENSECAP00000014722 | 95.5782 | 94.2177 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 93.8775 | 91.1565 | canis_lupus_familiaris | ENSCAFP00845005094 | 68.8279 | 66.8329 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.2177 | 93.1973 | panthera_pardus | ENSPPRP00000013031 | 99.6403 | 98.5611 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 93.3673 | 92.3469 | marmota_marmota_marmota | ENSMMMP00000021682 | 98.5637 | 97.4865 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.898 | 93.3673 | balaenoptera_musculus | ENSBMSP00010002067 | 99.4652 | 97.861 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 92.8571 | 91.1565 | cavia_porcellus | ENSCPOP00000018464 | 97.5 | 95.7143 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.2381 | 93.5374 | felis_catus | ENSFCAP00000028781 | 79.3201 | 77.9037 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.5782 | 94.0476 | ursus_americanus | ENSUAMP00000012092 | 95.416 | 93.8879 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 79.7619 | 76.7007 | monodelphis_domestica | ENSMODP00000002441 | 95.7143 | 92.0408 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.2177 | 93.0272 | bison_bison_bison | ENSBBBP00000015251 | 98.7522 | 97.5045 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.068 | 93.5374 | monodon_monoceros | ENSMMNP00015008856 | 99.6435 | 98.0392 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 93.7075 | 92.1769 | sus_scrofa | ENSSSCP00000001975 | 97.3498 | 95.7597 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 97.1088 | 95.4082 | microcebus_murinus | ENSMICP00000025915 | 97.1088 | 95.4082 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 70.2381 | 64.7959 | octodon_degus | ENSODEP00000000806 | 78.6667 | 72.5714 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 91.4966 | 89.7959 | octodon_degus | ENSODEP00000020668 | 96.4158 | 94.6237 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 93.0272 | 91.1565 | octodon_degus | ENSODEP00000021449 | 98.0287 | 96.0574 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 92.6871 | 91.6667 | jaculus_jaculus | ENSJJAP00000011856 | 96.6312 | 95.5674 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.5578 | 93.1973 | ovis_aries_rambouillet | ENSOARP00020024881 | 99.2857 | 97.8571 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.0476 | 92.8571 | ursus_maritimus | ENSUMAP00000013680 | 97.0175 | 95.7895 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 59.6939 | 57.6531 | ochotona_princeps | ENSOPRP00000016157 | 87.75 | 84.75 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.5578 | 93.1973 | delphinapterus_leucas | ENSDLEP00000010231 | 95.3688 | 93.9966 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.068 | 93.7075 | physeter_catodon | ENSPCTP00005031125 | 99.6435 | 98.2175 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 90.8163 | 89.6258 | mus_caroli | MGP_CAROLIEiJ_P0080460 | 95.0178 | 93.7722 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 90.6463 | 89.4558 | mus_musculus | ENSMUSP00000137079 | 94.8399 | 93.5943 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 89.6258 | 87.415 | dipodomys_ordii | ENSDORP00000008487 | 93.7722 | 91.4591 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.5578 | 93.0272 | mustela_putorius_furo | ENSMPUP00000015460 | 92.3588 | 90.8638 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 93.1973 | 90.9864 | aotus_nancymaae | ENSANAP00000019631 | 88.3871 | 86.2903 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.4082 | 93.7075 | saimiri_boliviensis_boliviensis | ENSSBOP00000016904 | 91.6667 | 90.0327 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 69.3878 | 68.8775 | vicugna_pacos | ENSVPAP00000005574 | 77.2727 | 76.7045 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 82.6531 | 82.3129 | chlorocebus_sabaeus | ENSCSAP00000013316 | 100 | 99.5885 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 90.3061 | 88.9456 | mesocricetus_auratus | ENSMAUP00000019520 | 96.3702 | 94.9183 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.068 | 93.5374 | phocoena_sinus | ENSPSNP00000000801 | 99.6435 | 98.0392 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 88.6054 | 85.8844 | camelus_dromedarius | ENSCDRP00005020699 | 74.2165 | 71.9373 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.0476 | 93.1973 | carlito_syrichta | ENSTSYP00000029316 | 97.7032 | 96.8198 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 82.483 | 81.4626 | panthera_tigris_altaica | ENSPTIP00000009231 | 98.778 | 97.556 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 91.1565 | 88.9456 | rattus_norvegicus | ENSRNOP00000073393 | 90.2357 | 88.0471 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 93.8775 | 92.6871 | prolemur_simus | ENSPSMP00000002713 | 97.1831 | 95.9507 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 88.6054 | 86.7347 | microtus_ochrogaster | ENSMOCP00000007164 | 93.0357 | 91.0714 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 90.6463 | 89.1156 | peromyscus_maniculatus_bairdii | ENSPEMP00000004476 | 95.0089 | 93.4046 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.2381 | 93.5374 | bos_mutus | ENSBMUP00000008923 | 93.0233 | 91.3621 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 91.1565 | 89.7959 | mus_spicilegus | ENSMSIP00000001837 | 68.0203 | 67.0051 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 90.9864 | 89.6258 | cricetulus_griseus_chok1gshd | ENSCGRP00001000460 | 95.1957 | 93.7722 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 77.381 | 75.3401 | pteropus_vampyrus | ENSPVAP00000011333 | 80.8171 | 78.6856 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 92.8571 | 90.1361 | myotis_lucifugus | ENSMLUP00000020563 | 94.6274 | 91.8544 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 79.7619 | 77.2109 | vombatus_ursinus | ENSVURP00010020701 | 95.7143 | 92.6531 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 93.0272 | 91.3265 | rhinolophus_ferrumequinum | ENSRFEP00010029781 | 97.6786 | 95.8929 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 93.1973 | 92.0068 | neovison_vison | ENSNVIP00000020124 | 97.6827 | 96.4349 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.2381 | 93.3673 | bos_taurus | ENSBTAP00000000819 | 92.869 | 91.0448 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 38.7755 | 37.2449 | heterocephalus_glaber_female | ENSHGLP00000026669 | 92.3077 | 88.664 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 93.8775 | 92.1769 | heterocephalus_glaber_female | ENSHGLP00000010198 | 97.6991 | 95.9292 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 51.3605 | 48.8095 | heterocephalus_glaber_female | ENSHGLP00000012786 | 75.3117 | 71.5711 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.7483 | 93.8775 | rhinopithecus_bieti | ENSRBIP00000001862 | 91.8434 | 90.0489 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.7279 | 91.6667 | canis_lupus_dingo | ENSCAFP00020020237 | 91.0131 | 88.0719 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.7279 | 93.0272 | sciurus_vulgaris | ENSSVLP00005010709 | 99.4643 | 97.6786 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 90.4762 | 86.5646 | phascolarctos_cinereus | ENSPCIP00000000174 | 95 | 90.8929 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 61.9048 | 59.1837 | procavia_capensis | ENSPCAP00000008999 | 68.8091 | 65.7845 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 93.5374 | 92.1769 | nannospalax_galili | ENSNGAP00000009277 | 97.8648 | 96.4413 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 90.9864 | 88.4354 | bos_grunniens | ENSBGRP00000020158 | 80.5723 | 78.3133 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 93.0272 | 92.0068 | chinchilla_lanigera | ENSCLAP00000013892 | 98.5586 | 97.4775 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.2381 | 93.3673 | cervus_hanglu_yarkandensis | ENSCHYP00000033490 | 93.0233 | 91.196 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 93.3673 | 91.8367 | catagonus_wagneri | ENSCWAP00000018901 | 97.861 | 96.2567 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.7279 | 93.5374 | ictidomys_tridecemlineatus | ENSSTOP00000017144 | 98.4099 | 97.1731 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.0476 | 93.1973 | otolemur_garnettii | ENSOGAP00000021502 | 97.5309 | 96.649 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.2381 | 93.5374 | cebus_imitator | ENSCCAP00000039214 | 91.5033 | 89.8693 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.2177 | 93.0272 | urocitellus_parryii | ENSUPAP00010018108 | 98.4014 | 97.1581 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 94.898 | 93.3673 | moschus_moschiferus | ENSMMSP00000030984 | 92.691 | 91.196 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 95.7483 | 93.8775 | rhinopithecus_roxellana | ENSRROP00000024454 | 91.8434 | 90.0489 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 90.6463 | 89.2857 | mus_pahari | MGP_PahariEiJ_P0017311 | 94.8399 | 93.4164 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 87.7551 | 85.034 | propithecus_coquereli | ENSPCOP00000011019 | 87.6061 | 84.8896 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 46.5986 | 28.7415 | caenorhabditis_elegans | B0414.5.1 | 36.7785 | 22.6846 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 54.5918 | 36.3946 | drosophila_melanogaster | FBpp0306565 | 35.082 | 23.388 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 46.5986 | 36.3946 | ciona_intestinalis | ENSCINP00000003938 | 67.6543 | 52.8395 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 55.102 | 46.4286 | petromyzon_marinus | ENSPMAP00000009770 | 81 | 68.25 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 55.7823 | 42.0068 | eptatretus_burgeri | ENSEBUP00000001466 | 66.5314 | 50.1014 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 60.3741 | 46.9388 | eptatretus_burgeri | ENSEBUP00000024903 | 53.3033 | 41.4414 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 19.2177 | 13.7755 | eptatretus_burgeri | ENSEBUP00000027201 | 51.1312 | 36.6516 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 44.898 | 31.8027 | eptatretus_burgeri | ENSEBUP00000014135 | 54.433 | 38.5567 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 71.5986 | 63.0952 | callorhinchus_milii | ENSCMIP00000010555 | 84.7083 | 74.6479 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 70.068 | 64.966 | latimeria_chalumnae | ENSLACP00000018771 | 84.7737 | 78.6008 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 81.8027 | 71.0884 | lepisosteus_oculatus | ENSLOCP00000018477 | 85.1327 | 73.9823 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 76.1905 | 65.6463 | clupea_harengus | ENSCHAP00000048076 | 78.3217 | 67.4825 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 73.8095 | 62.2449 | danio_rerio | ENSDARP00000089674 | 77.6386 | 65.4741 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 59.5238 | 47.2789 | danio_rerio | ENSDARP00000095525 | 67.0498 | 53.2567 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.8299 | 62.2449 | carassius_auratus | ENSCARP00000027823 | 78.0142 | 64.8936 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 59.0136 | 47.7891 | carassius_auratus | ENSCARP00000124589 | 68.9861 | 55.8648 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 51.0204 | 40.4762 | carassius_auratus | ENSCARP00000056109 | 67.2646 | 53.3632 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 56.8027 | 45.2381 | carassius_auratus | ENSCARP00000093022 | 59.8566 | 47.6702 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 75.1701 | 63.0952 | carassius_auratus | ENSCARP00000030711 | 76.6031 | 64.2981 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 58.1633 | 47.7891 | carassius_auratus | ENSCARP00000106155 | 69.9387 | 57.4642 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 56.8027 | 44.5578 | carassius_auratus | ENSCARP00000109683 | 60.9489 | 47.8102 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 58.8435 | 48.1293 | carassius_auratus | ENSCARP00000031054 | 68.7873 | 56.2624 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 68.8775 | 56.2925 | astyanax_mexicanus | ENSAMXP00000031387 | 72.5806 | 59.319 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 77.2109 | 66.1565 | astyanax_mexicanus | ENSAMXP00000004933 | 79.7891 | 68.3656 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 77.381 | 67.1769 | ictalurus_punctatus | ENSIPUP00000031096 | 81.8345 | 71.0432 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 62.0748 | 48.6395 | ictalurus_punctatus | ENSIPUP00000019993 | 69.3916 | 54.3726 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 70.5782 | 60.5442 | esox_lucius | ENSELUP00000042735 | 72.2997 | 62.0209 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 77.7211 | 67.6871 | esox_lucius | ENSELUP00000075925 | 76.0399 | 66.223 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 64.6258 | 53.4014 | electrophorus_electricus | ENSEEEP00000037743 | 66.7838 | 55.1845 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 76.0204 | 64.7959 | electrophorus_electricus | ENSEEEP00000042816 | 80.5405 | 68.6487 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 76.5306 | 66.6667 | oncorhynchus_kisutch | ENSOKIP00005110351 | 76.9231 | 67.0085 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 71.4286 | 61.5646 | oncorhynchus_kisutch | ENSOKIP00005100366 | 70.5882 | 60.8403 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 76.3605 | 66.3265 | oncorhynchus_mykiss | ENSOMYP00000131494 | 76.7521 | 66.6667 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 71.7687 | 62.2449 | oncorhynchus_mykiss | ENSOMYP00000065970 | 70.9244 | 61.5126 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 77.0408 | 66.8367 | oncorhynchus_mykiss | ENSOMYP00000131555 | 75.8794 | 65.8291 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 75.3401 | 65.3061 | salmo_salar | ENSSSAP00000095645 | 77.7193 | 67.3684 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 77.0408 | 66.4966 | salmo_salar | ENSSSAP00000107995 | 77.7015 | 67.0669 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 69.2177 | 60.034 | salmo_salar | ENSSSAP00000069205 | 61.6667 | 53.4848 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 70.2381 | 61.0544 | salmo_trutta | ENSSTUP00000074637 | 72.0768 | 62.6527 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 78.4014 | 67.8571 | salmo_trutta | ENSSTUP00000067071 | 75.9473 | 65.7331 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 76.5306 | 66.3265 | salmo_trutta | ENSSTUP00000097273 | 76.661 | 66.4395 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 76.8708 | 65.1361 | gadus_morhua | ENSGMOP00000009095 | 75.9664 | 64.3698 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.3197 | 61.9048 | fundulus_heteroclitus | ENSFHEP00000029843 | 75.7366 | 63.0849 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 69.3878 | 55.7823 | poecilia_reticulata | ENSPREP00000008807 | 66.9951 | 53.8588 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 70.068 | 57.9932 | xiphophorus_maculatus | ENSXMAP00000039719 | 69.8305 | 57.7966 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 75.1701 | 62.585 | oryzias_latipes | ENSORLP00000029310 | 75.6849 | 63.0137 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 76.5306 | 63.6054 | cyclopterus_lumpus | ENSCLMP00005001779 | 71.5421 | 59.4595 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.4898 | 62.0748 | oreochromis_niloticus | ENSONIP00000007263 | 75.9099 | 63.2582 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.4898 | 62.0748 | haplochromis_burtoni | ENSHBUP00000033074 | 76.5734 | 63.8112 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 67.517 | 56.8027 | astatotilapia_calliptera | ENSACLP00000018777 | 79.4 | 66.8 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.3197 | 62.585 | sparus_aurata | ENSSAUP00010008843 | 75.7366 | 63.7782 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 75.3401 | 62.585 | lates_calcarifer | ENSLCAP00010041066 | 76.6436 | 63.6678 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 84.1837 | 76.5306 | xenopus_tropicalis | ENSXETP00000078803 | 85.6401 | 77.8547 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 88.6054 | 82.8231 | chrysemys_picta_bellii | ENSCPBP00000007879 | 92.8699 | 86.8093 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 85.8844 | 79.932 | sphenodon_punctatus | ENSSPUP00000008149 | 87.9791 | 81.8815 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 87.7551 | 82.6531 | crocodylus_porosus | ENSCPRP00005007712 | 90.6854 | 85.413 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 68.3673 | 60.034 | laticauda_laticaudata | ENSLLTP00000012552 | 72.1724 | 63.3752 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 78.9116 | 71.2585 | notechis_scutatus | ENSNSUP00000027205 | 86.5672 | 78.1716 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 71.9388 | 65.3061 | pseudonaja_textilis | ENSPTXP00000019200 | 87.9418 | 79.8337 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 81.8027 | 76.1905 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000017038 | 90.2439 | 84.0525 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 87.0748 | 80.6122 | gallus_gallus | ENSGALP00010034633 | 90.4594 | 83.7456 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 87.585 | 80.9524 | meleagris_gallopavo | ENSMGAP00000022818 | 91.4742 | 84.5471 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 81.2925 | 75.1701 | serinus_canaria | ENSSCAP00000022144 | 88.8476 | 82.1561 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 86.5646 | 80.7823 | parus_major | ENSPMJP00000009097 | 91.3824 | 85.2783 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 73.8095 | 66.8367 | anolis_carolinensis | ENSACAP00000020487 | 79.9263 | 72.3757 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 69.0476 | 56.4626 | neolamprologus_brichardi | ENSNBRP00000026227 | 71.7314 | 58.6572 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 71.9388 | 64.966 | naja_naja | ENSNNAP00000018315 | 87.9418 | 79.4179 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 78.2313 | 71.5986 | podarcis_muralis | ENSPMRP00000023563 | 82.7338 | 75.7194 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 86.5646 | 80.7823 | geospiza_fortis | ENSGFOP00000000273 | 88.986 | 83.042 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 82.3129 | 75.1701 | taeniopygia_guttata | ENSTGUP00000027993 | 82.0339 | 74.9153 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 49.6599 | 44.3878 | erpetoichthys_calabaricus | ENSECRP00000032497 | 81.5642 | 72.905 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 70.7483 | 59.3537 | kryptolebias_marmoratus | ENSKMAP00000002576 | 73.3686 | 61.552 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 79.932 | 73.6395 | salvator_merianae | ENSSMRP00000028993 | 87.8505 | 80.9346 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 73.8095 | 61.9048 | oryzias_javanicus | ENSOJAP00000001782 | 75.3472 | 63.1944 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 65.9864 | 56.2925 | amphilophus_citrinellus | ENSACIP00000017538 | 80.1653 | 68.3884 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.8299 | 62.2449 | dicentrarchus_labrax | ENSDLAP00005050915 | 75.4717 | 62.7787 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 72.9592 | 61.7347 | amphiprion_percula | ENSAPEP00000002668 | 70.6755 | 59.8023 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 20.7483 | 15.9864 | oryzias_sinensis | ENSOSIP00000003652 | 68.9266 | 53.1073 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 49.6599 | 39.1156 | oryzias_sinensis | ENSOSIP00000038973 | 69.5238 | 54.7619 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 54.7619 | 52.381 | pelodiscus_sinensis | ENSPSIP00000018807 | 96.1194 | 91.9403 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 88.0952 | 81.9728 | gopherus_evgoodei | ENSGEVP00005023256 | 85.4785 | 79.538 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 82.9932 | 78.2313 | chelonoidis_abingdonii | ENSCABP00000029342 | 93.4866 | 88.1226 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.1497 | 61.2245 | seriola_dumerili | ENSSDUP00000000936 | 73.0318 | 60.3015 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 76.0204 | 62.7551 | cottoperca_gobio | ENSCGOP00000010212 | 69.3023 | 57.2093 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 88.2653 | 82.3129 | struthio_camelus_australis | ENSSCUP00000006795 | 81.6038 | 76.1006 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 87.7551 | 81.6327 | anser_brachyrhynchus | ENSABRP00000019904 | 91.9786 | 85.5615 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 66.6667 | 54.4218 | gasterosteus_aculeatus | ENSGACP00000023090 | 73.4082 | 59.9251 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 71.5986 | 58.3333 | cynoglossus_semilaevis | ENSCSEP00000013408 | 70.1667 | 57.1667 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.3197 | 62.415 | poecilia_formosa | ENSPFOP00000016110 | 75.6055 | 63.4948 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 75.3401 | 62.7551 | myripristis_murdjan | ENSMMDP00005050218 | 75.3401 | 62.7551 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 75.5102 | 62.9252 | sander_lucioperca | ENSSLUP00000037078 | 71.4976 | 59.5813 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 87.2449 | 81.4626 | strigops_habroptila | ENSSHBP00005016657 | 91.4438 | 85.3832 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.6599 | 62.585 | amphiprion_ocellaris | ENSAOCP00000020077 | 76.6143 | 64.2234 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 88.4354 | 82.483 | terrapene_carolina_triunguis | ENSTMTP00000021707 | 90.7504 | 84.6422 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 70.2381 | 60.7143 | hucho_hucho | ENSHHUP00000004489 | 74.5487 | 64.4404 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 72.1088 | 62.0748 | hucho_hucho | ENSHHUP00000067189 | 67.84 | 58.4 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.6599 | 64.6258 | hucho_hucho | ENSHHUP00000075714 | 76.2153 | 65.9722 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 71.0884 | 58.5034 | betta_splendens | ENSBSLP00000025976 | 70.489 | 58.0101 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 77.0408 | 66.6667 | pygocentrus_nattereri | ENSPNAP00000011105 | 80.4618 | 69.627 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 66.3265 | 54.5918 | pygocentrus_nattereri | ENSPNAP00000020686 | 69.7674 | 57.424 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.6599 | 62.415 | anabas_testudineus | ENSATEP00000029385 | 74.4068 | 62.2034 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 21.4286 | 15.4762 | ciona_savignyi | ENSCSAVP00000004444 | 52.0661 | 37.6033 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 70.7483 | 58.8435 | seriola_lalandi_dorsalis | ENSSLDP00000029891 | 77.037 | 64.0741 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 71.7687 | 60.5442 | labrus_bergylta | ENSLBEP00000022627 | 76.311 | 64.3761 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 71.7687 | 58.8435 | pundamilia_nyererei | ENSPNYP00000014250 | 74.8227 | 61.3475 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.1497 | 61.3946 | mastacembelus_armatus | ENSMAMP00000038374 | 77.305 | 64.0071 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.8299 | 62.9252 | stegastes_partitus | ENSSPAP00000029303 | 73.8255 | 62.0805 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 70.2381 | 59.1837 | acanthochromis_polyacanthus | ENSAPOP00000025034 | 74.8188 | 63.0435 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 88.2653 | 82.483 | aquila_chrysaetos_chrysaetos | ENSACCP00020002787 | 88.8699 | 83.0479 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.3197 | 62.0748 | nothobranchius_furzeri | ENSNFUP00015024985 | 75.6055 | 63.1488 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 67.1769 | 57.1429 | cyprinodon_variegatus | ENSCVAP00000022299 | 79.6371 | 67.7419 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 77.2109 | 68.3673 | denticeps_clupeoides | ENSDCDP00000035098 | 72.7564 | 64.4231 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.1497 | 63.2653 | denticeps_clupeoides | ENSDCDP00000011982 | 76.8959 | 65.6085 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 75.3401 | 63.4354 | larimichthys_crocea | ENSLCRP00005035886 | 76.1168 | 64.0893 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 73.9796 | 63.2653 | takifugu_rubripes | ENSTRUP00000042511 | 76.049 | 65.035 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 86.9048 | 80.4422 | ficedula_albicollis | ENSFALP00000017906 | 86.9048 | 80.4422 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 73.6395 | 61.5646 | hippocampus_comes | ENSHCOP00000014899 | 75.4355 | 63.0662 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 54.4218 | 42.3469 | cyprinus_carpio_carpio | ENSCCRP00000081236 | 62.8684 | 48.9194 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 60.034 | 48.2993 | cyprinus_carpio_carpio | ENSCCRP00000132008 | 66.1049 | 53.1835 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 59.0136 | 48.1293 | cyprinus_carpio_carpio | ENSCCRP00000174774 | 69.5391 | 56.7134 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.1497 | 62.9252 | cyprinus_carpio_carpio | ENSCCRP00000148343 | 75.5633 | 64.1248 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 73.6395 | 62.415 | cyprinus_carpio_carpio | ENSCCRP00000120216 | 75.5672 | 64.0489 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 87.585 | 80.7823 | coturnix_japonica | ENSCJPP00005001533 | 87.1404 | 80.3723 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 73.4694 | 62.2449 | poecilia_latipinna | ENSPLAP00000023460 | 74.7405 | 63.3218 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 44.5578 | 35.2041 | sinocyclocheilus_grahami | ENSSGRP00000085207 | 63.4383 | 50.1211 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 58.6735 | 47.2789 | sinocyclocheilus_grahami | ENSSGRP00000075296 | 68.5885 | 55.2684 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 63.2653 | 53.5714 | sinocyclocheilus_grahami | ENSSGRP00000101099 | 77.6618 | 65.762 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 53.9116 | 45.068 | sinocyclocheilus_grahami | ENSSGRP00000007817 | 75.1185 | 62.7962 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.3197 | 61.9048 | maylandia_zebra | ENSMZEP00005018269 | 75.7366 | 63.0849 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 74.3197 | 62.415 | tetraodon_nigroviridis | ENSTNIP00000012445 | 74.8288 | 62.8425 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 80.4422 | 68.0272 | paramormyrops_kingsleyae | ENSPKIP00000007417 | 79.8987 | 67.5676 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 55.9524 | 47.7891 | paramormyrops_kingsleyae | ENSPKIP00000032099 | 72.1491 | 61.6228 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 79.2517 | 68.5374 | scleropages_formosus | ENSSFOP00015070875 | 80.7626 | 69.844 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 69.5578 | 60.2041 | scleropages_formosus | ENSSFOP00015069301 | 75.0459 | 64.9541 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 83.8435 | 76.3605 | leptobrachium_leishanense | ENSLLEP00000022979 | 83.9864 | 76.4906 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 75.1701 | 61.5646 | scophthalmus_maximus | ENSSMAP00000037899 | 71.2903 | 58.3871 |
| ENSG00000214575 | homo_sapiens | ENSP00000507835 | 71.4286 | 59.1837 | oryzias_melastigma | ENSOMEP00000005555 | 71.4286 | 59.1837 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000900 | enables mRNA regulatory element binding translation repressor activity | 2 | IBA,ISS | Homo_sapiens(9606) | Function |
| GO:0000932 | located_in P-body | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0003730 | enables mRNA 3-UTR binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 2 | IBA,ISS | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 3 | IBA,IDA,ISS | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0006397 | involved_in mRNA processing | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006412 | involved_in translation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008135 | enables translation factor activity, RNA binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0014069 | located_in postsynaptic density | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0016020 | located_in membrane | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0030425 | located_in dendrite | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0032869 | involved_in cellular response to insulin stimulus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0035925 | enables mRNA 3-UTR AU-rich region binding | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0043005 | is_active_in neuron projection | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0043022 | enables ribosome binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0045202 | is_active_in synapse | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0046872 | enables metal ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0071230 | involved_in cellular response to amino acid stimulus | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0071456 | involved_in cellular response to hypoxia | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1990124 | part_of messenger ribonucleoprotein complex | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:2000766 | involved_in negative regulation of cytoplasmic translation | 2 | IBA,IMP | Homo_sapiens(9606) | Process |