Welcome to
RBP World!
Functions and Diseases
| RBP Type | Non-canonical_RBPs |
| Diseases | Cancer |
| Drug | N.A. |
| Main interacting RNAs | N.A. |
| Moonlighting functions | Transmemebrane |
| Localizations | N.A. |
| BulkPerturb-seq | N.A. |
Description
| Ensembl ID | ENSG00000206503 | Gene ID | 3105 | Accession | 4931 |
| Symbol | HLA-A | Alias | HLAA | Full Name | major histocompatibility complex, class I, A |
| Status | Confidence | Length | 8313 bases | Strand | Plus strand |
| Position | 6 : 29941260 - 29949572 | RNA binding domain | N.A. | ||
| Summary | HLA-A belongs to the HLA class I heavy chain paralogues. This class I molecule is a heterodimer consisting of a heavy chain and a light chain (beta-2 microglobulin). The heavy chain is anchored in the membrane. Class I molecules play a central role in the immune system by presenting peptides derived from the endoplasmic reticulum lumen so that they can be recognized by cytotoxic T cells. They are expressed in nearly all cells. The heavy chain is approximately 45 kDa and its gene contains 8 exons. Exon 1 encodes the leader peptide, exons 2 and 3 encode the alpha1 and alpha2 domains, which both bind the peptide, exon 4 encodes the alpha3 domain, exon 5 encodes the transmembrane region, and exons 6 and 7 encode the cytoplasmic tail. Polymorphisms within exon 2 and exon 3 are responsible for the peptide binding specificity of each class one molecule. Typing for these polymorphisms is routinely done for bone marrow and kidney transplantation. More than 6000 HLA-A alleles have been described. The HLA system plays an important role in the occurrence and outcome of infectious diseases, including those caused by the malaria parasite, the human immunodeficiency virus (HIV), and the severe acute respiratory syndrome coronavirus (SARS-CoV). The structural spike and the nucleocapsid proteins of the novel coronavirus SARS-CoV-2, which causes coronavirus disease 2019 (COVID-19), are reported to contain multiple Class I epitopes with predicted HLA restrictions. Individual HLA genetic variation may help explain different immune responses to a virus across a population.[provided by RefSeq, Aug 2020] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 29496759 | The RNA-binding Protein MEX3B Mediates Resistance to Cancer Immunotherapy by Downregulating HLA-A Expression. | Lu Huang | 2018-07-15 | Clinical cancer research : an official journal of the American Association for Cancer Research |
| 34353849 | Deregulation of HLA-I in cancer and its central importance for immunotherapy. | Ahmet Hazini | 2021-08-01 | Journal for immunotherapy of cancer |
| 20512327 | Peptide microarrays for the profiling of cytotoxic T-lymphocyte activity using minimum numbers of cells. | Antje Hoff | 2010-09-01 | Cancer immunology, immunotherapy : CII |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 58224 | Respiratory Function Tests | 2.0090000E-006 | 20010835 |
| 58225 | Respiratory Function Tests | 3.2550000E-006 | 20010835 |
| 58226 | Respiratory Function Tests | 1.9450000E-006 | 20010835 |
| 64797 | Multiple Sclerosis | 1E-11 | 22190364 |
| 76196 | beta 2-Microglobulin | 2E-22 | 23417110 |
| 78076 | Menarche | 3E-10 | 25231870 |
| 93727 | Anticonvulsants | 1E-7 | 21428769 |
| 97602 | Nasopharyngeal Neoplasms | 4E-20 | 19664746 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
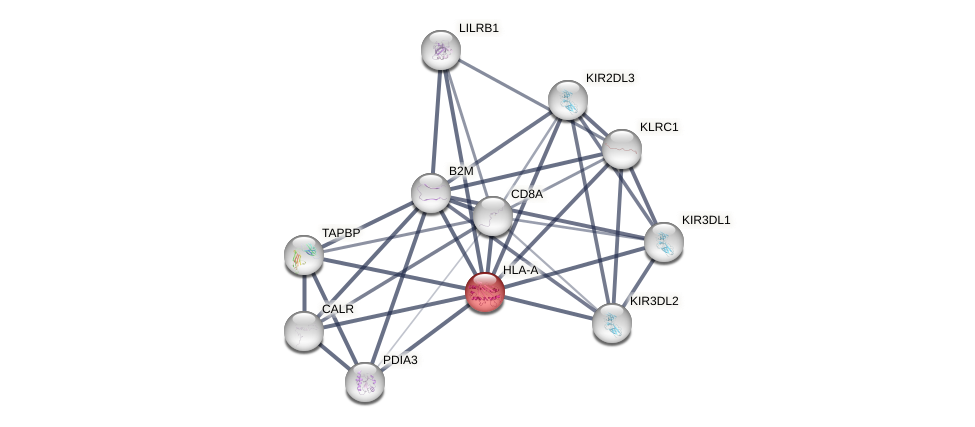
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000206503 | homo_sapiens | ENSP00000365402 | 87.9781 | 81.1475 | homo_sapiens | ENSP00000366005 | 88.2192 | 81.3699 |
| ENSG00000206503 | homo_sapiens | ENSP00000399168 | 90.6077 | 85.3591 | homo_sapiens | ENSP00000366005 | 89.863 | 84.6575 |
| ENSG00000206503 | homo_sapiens | ENSP00000259951 | 69.2308 | 62.6697 | homo_sapiens | ENSP00000366005 | 83.8356 | 75.8904 |
| ENSG00000206503 | homo_sapiens | ENSP00000353472 | 90.2367 | 82.2485 | homo_sapiens | ENSP00000366005 | 83.5616 | 76.1644 |
| ENSG00000206503 | homo_sapiens | ENSP00000365817 | 87.7095 | 77.6536 | homo_sapiens | ENSP00000366005 | 86.0274 | 76.1644 |
| ENSG00000206503 | homo_sapiens | ENSP00000221466 | 42.7397 | 25.7534 | homo_sapiens | ENSP00000366005 | 42.7397 | 25.7534 |
| ENSG00000206503 | homo_sapiens | ENSP00000357150 | 40.2402 | 21.021 | homo_sapiens | ENSP00000366005 | 36.7123 | 19.1781 |
| ENSG00000206503 | homo_sapiens | ENSP00000289429 | 41.896 | 24.4648 | homo_sapiens | ENSP00000366005 | 37.5342 | 21.9178 |
| ENSG00000206503 | homo_sapiens | ENSP00000357149 | 35.3093 | 20.1031 | homo_sapiens | ENSP00000366005 | 37.5342 | 21.3699 |
| ENSG00000206503 | homo_sapiens | ENSP00000357152 | 39.9399 | 21.021 | homo_sapiens | ENSP00000366005 | 36.4384 | 19.1781 |
| ENSG00000206503 | homo_sapiens | ENSP00000501100 | 37.6119 | 20.597 | homo_sapiens | ENSP00000366005 | 34.5205 | 18.9041 |
| ENSG00000206503 | homo_sapiens | ENSP00000356320 | 39.8374 | 23.9837 | homo_sapiens | ENSP00000366005 | 26.8493 | 16.1644 |
| ENSG00000206503 | homo_sapiens | ENSP00000356310 | 39.8374 | 23.9837 | homo_sapiens | ENSP00000366005 | 26.8493 | 16.1644 |
| ENSG00000206503 | homo_sapiens | ENSP00000417404 | 54.8851 | 37.931 | homo_sapiens | ENSP00000366005 | 52.3288 | 36.1644 |
| ENSG00000206503 | homo_sapiens | ENSP00000356308 | 40.5738 | 25 | homo_sapiens | ENSP00000366005 | 27.1233 | 16.7123 |
| ENSG00000206503 | homo_sapiens | ENSP00000356552 | 54.2522 | 35.7771 | homo_sapiens | ENSP00000366005 | 50.6849 | 33.4247 |
| ENSG00000206503 | homo_sapiens | ENSP00000349709 | 37.2624 | 19.3916 | homo_sapiens | ENSP00000366005 | 26.8493 | 13.9726 |
| ENSG00000206503 | homo_sapiens | ENSP00000292401 | 52.0134 | 36.5772 | homo_sapiens | ENSP00000366005 | 42.4658 | 29.863 |
| ENSG00000206503 | homo_sapiens | ENSP00000229708 | 41.8033 | 24.5902 | homo_sapiens | ENSP00000366005 | 27.9452 | 16.4384 |
| ENSG00000206503 | homo_sapiens | ENSP00000356329 | 30.2395 | 18.5629 | homo_sapiens | ENSP00000366005 | 27.6712 | 16.9863 |
| ENSG00000206503 | homo_sapiens | ENSP00000252229 | 42.5587 | 26.3708 | homo_sapiens | ENSP00000366005 | 44.6575 | 27.6712 |
| ENSG00000206503 | homo_sapiens | ENSP00000413079 | 43.3735 | 27.4096 | homo_sapiens | ENSP00000366005 | 39.4521 | 24.9315 |
| ENSG00000206503 | homo_sapiens | ENSP00000448516 | 89.8477 | 88.5787 | homo_sapiens | ENSP00000366005 | 96.9863 | 95.6164 |
| ENSG00000206503 | homo_sapiens | ENSP00000408986 | 96.1644 | 93.4247 | homo_sapiens | ENSP00000366005 | 96.1644 | 93.4247 |
| ENSG00000206503 | homo_sapiens | ENSP00000388526 | 93.8005 | 92.1833 | homo_sapiens | ENSP00000366005 | 95.3425 | 93.6986 |
| ENSG00000206503 | homo_sapiens | ENSP00000449453 | 89.8477 | 88.5787 | homo_sapiens | ENSP00000366005 | 96.9863 | 95.6164 |
| ENSG00000206503 | homo_sapiens | ENSP00000416233 | 93.531 | 91.9137 | homo_sapiens | ENSP00000366005 | 95.0685 | 93.4247 |
| ENSG00000206503 | homo_sapiens | ENSP00000483385 | 88.5787 | 85.533 | homo_sapiens | ENSP00000366005 | 95.6164 | 92.3288 |
| ENSG00000206503 | homo_sapiens | ENSP00000410645 | 94.2466 | 91.5069 | homo_sapiens | ENSP00000366005 | 94.2466 | 91.5069 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 94.2466 | 90.411 | pongo_abelii | ENSPPYP00000018330 | 93.2249 | 89.4309 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 95.8904 | 92.3288 | pongo_abelii | ENSPPYP00000036080 | 95.8904 | 92.3288 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 97.5342 | 96.1644 | pan_troglodytes | ENSPTRP00000089515 | 97.5342 | 96.1644 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 93.4247 | 89.3151 | pan_troglodytes | ENSPTRP00000068372 | 93.4247 | 89.3151 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 60.274 | 46.3014 | ornithorhynchus_anatinus | ENSOANP00000039984 | 51.1628 | 39.3023 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 27.3973 | 22.1918 | ornithorhynchus_anatinus | ENSOANP00000028190 | 45.045 | 36.4865 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 47.9452 | 32.3288 | ornithorhynchus_anatinus | ENSOANP00000002884 | 52.8701 | 35.6495 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 56.9863 | 42.7397 | ornithorhynchus_anatinus | ENSOANP00000008299 | 64 | 48 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 37.2603 | 29.589 | ornithorhynchus_anatinus | ENSOANP00000042132 | 51.1278 | 40.6015 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 58.9041 | 46.3014 | ornithorhynchus_anatinus | ENSOANP00000049233 | 45.8422 | 36.0341 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 55.3425 | 43.5616 | ornithorhynchus_anatinus | ENSOANP00000051511 | 59.587 | 46.9027 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 55.0685 | 41.3699 | ornithorhynchus_anatinus | ENSOANP00000002957 | 52.3438 | 39.3229 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 27.3973 | 20 | ornithorhynchus_anatinus | ENSOANP00000004195 | 57.8035 | 42.1965 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 71.7808 | 59.4521 | sarcophilus_harrisii | ENSSHAP00000012590 | 72.5762 | 60.1108 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 66.8493 | 53.1507 | sarcophilus_harrisii | ENSSHAP00000023414 | 68.5393 | 54.4944 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.8767 | 60.8219 | sarcophilus_harrisii | ENSSHAP00000035869 | 73.6842 | 61.4958 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 62.4658 | 48.7671 | sarcophilus_harrisii | ENSSHAP00000002229 | 63.1579 | 49.3075 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.6027 | 61.0959 | sarcophilus_harrisii | ENSSHAP00000029759 | 73.4072 | 61.7729 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.3288 | 61.0959 | sarcophilus_harrisii | ENSSHAP00000012589 | 73.1302 | 61.7729 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 44.1096 | 34.5205 | notamacropus_eugenii | ENSMEUP00000007792 | 65.9836 | 51.6393 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 55.6164 | 48.4931 | dasypus_novemcinctus | ENSDNOP00000025342 | 63.2399 | 55.1402 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 62.7397 | 52.8767 | erinaceus_europaeus | ENSEEUP00000012396 | 68.5629 | 57.7844 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 62.4658 | 51.7808 | erinaceus_europaeus | ENSEEUP00000012436 | 66.8622 | 55.4252 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 38.6301 | 35.3425 | tursiops_truncatus | ENSTTRP00000016042 | 67.7885 | 62.0192 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 71.2329 | 59.4521 | vulpes_vulpes | ENSVVUP00000018614 | 60.4651 | 50.4651 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 73.9726 | 64.1096 | vulpes_vulpes | ENSVVUP00000025400 | 65.2174 | 56.5217 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 44.3836 | 39.1781 | vulpes_vulpes | ENSVVUP00000020588 | 66.1225 | 58.3673 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.3288 | 64.3836 | vulpes_vulpes | ENSVVUP00000020574 | 73.1302 | 65.097 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.8767 | 64.3836 | vulpes_vulpes | ENSVVUP00000003408 | 59.2428 | 52.3385 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 43.5616 | 35.6164 | vulpes_vulpes | ENSVVUP00000037459 | 58.2418 | 47.619 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.274 | 71.5069 | equus_asinus | ENSEASP00005038186 | 49.4932 | 44.0878 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 74.7945 | 64.3836 | equus_asinus | ENSEASP00005062700 | 62.4714 | 53.7757 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.1781 | 69.863 | equus_asinus | ENSEASP00005035207 | 71.7122 | 63.2754 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 81.9178 | 75.0685 | equus_asinus | ENSEASP00005002182 | 86.1671 | 78.9625 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.6301 | 69.3151 | equus_asinus | ENSEASP00005025372 | 75.5263 | 66.5789 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 74.5205 | 63.2877 | equus_asinus | ENSEASP00005038099 | 56.3147 | 47.8261 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.3562 | 68.4931 | capra_hircus | ENSCHIP00000028898 | 79.2244 | 69.2521 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.726 | 71.5069 | capra_hircus | ENSCHIP00000003807 | 80.8333 | 72.5 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 64.9315 | 56.9863 | capra_hircus | ENSCHIP00000008860 | 78.2178 | 68.6469 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 71.7808 | 60.8219 | capra_hircus | ENSCHIP00000022962 | 70.0535 | 59.3583 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 73.9726 | 63.8356 | capra_hircus | ENSCHIP00000009976 | 70.6806 | 60.9948 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.726 | 69.3151 | capra_hircus | ENSCHIP00000007192 | 75.9791 | 66.0574 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 64.1096 | 56.9863 | capra_hircus | ENSCHIP00000018926 | 72.2222 | 64.1975 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.1781 | 69.863 | capra_hircus | ENSCHIP00000021018 | 80.9524 | 71.4286 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.0822 | 69.589 | capra_hircus | ENSCHIP00000005504 | 77.2358 | 68.8347 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 68.2192 | 58.9041 | capra_hircus | ENSCHIP00000020264 | 69.1667 | 59.7222 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.4658 | 73.4247 | capra_hircus | ENSCHIP00000024897 | 83.3795 | 74.2382 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 73.1507 | 65.4795 | capra_hircus | ENSCHIP00000024970 | 72.1622 | 64.5946 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 68.7671 | 60.5479 | capra_hircus | ENSCHIP00000029125 | 66.9333 | 58.9333 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 67.3973 | 59.4521 | capra_hircus | ENSCHIP00000000563 | 74.5455 | 65.7576 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.8219 | 71.7808 | capra_hircus | ENSCHIP00000021019 | 75.2551 | 66.8367 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 77.8082 | 69.863 | capra_hircus | ENSCHIP00000020123 | 81.6092 | 73.2759 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 59.1781 | 51.2329 | capra_hircus | ENSCHIP00000020161 | 60.8451 | 52.6761 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.0685 | 66.8493 | loxodonta_africana | ENSLAFP00000004650 | 81.0651 | 72.1893 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.0822 | 68.7671 | loxodonta_africana | ENSLAFP00000025493 | 77.4457 | 68.2065 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 69.0411 | 61.0959 | loxodonta_africana | ENSLAFP00000028223 | 81.8182 | 72.4026 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 53.4247 | 46.8493 | loxodonta_africana | ENSLAFP00000016344 | 82.2785 | 72.1519 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.0822 | 68.7671 | loxodonta_africana | ENSLAFP00000002478 | 83.8235 | 73.8235 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 81.3699 | 71.7808 | panthera_leo | ENSPLOP00000020921 | 82.0442 | 72.3757 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 58.6301 | 50.411 | panthera_leo | ENSPLOP00000024811 | 64.6526 | 55.5891 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 63.8356 | 57.2603 | panthera_leo | ENSPLOP00000019815 | 80.3448 | 72.069 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.6027 | 64.1096 | panthera_leo | ENSPLOP00000019364 | 65.5941 | 57.9208 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 46.8493 | 39.1781 | panthera_leo | ENSPLOP00000019626 | 70.9544 | 59.3361 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.8904 | 67.1233 | panthera_leo | ENSPLOP00000022888 | 76.3085 | 67.4931 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.4658 | 73.1507 | panthera_leo | ENSPLOP00000019589 | 73.7745 | 65.4412 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 76.7123 | 67.1233 | panthera_leo | ENSPLOP00000012188 | 69.1358 | 60.4938 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 49.0411 | 41.0959 | panthera_leo | ENSPLOP00000018916 | 49.3113 | 41.3223 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.9041 | 69.863 | ailuropoda_melanoleuca | ENSAMEP00000030207 | 79.1209 | 70.0549 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.1781 | 72.0548 | ailuropoda_melanoleuca | ENSAMEP00000028011 | 83.046 | 75.5747 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 55.8904 | 48.2192 | ailuropoda_melanoleuca | ENSAMEP00000032186 | 79.3774 | 68.4825 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.6164 | 68.4931 | ailuropoda_melanoleuca | ENSAMEP00000027689 | 70.9512 | 64.2673 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 69.3151 | 60 | ailuropoda_melanoleuca | ENSAMEP00000002421 | 78.5714 | 68.0124 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 60.8219 | 50.137 | bos_indicus_hybrid | ENSBIXP00005035551 | 47.234 | 38.9362 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 30.137 | 26.3014 | bos_indicus_hybrid | ENSBIXP00005009961 | 61.7978 | 53.9326 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 83.5616 | 76.4384 | bos_indicus_hybrid | ENSBIXP00005009909 | 75.1232 | 68.7192 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 70.6849 | 62.7397 | bos_indicus_hybrid | ENSBIXP00005033927 | 63.5468 | 56.4039 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.3425 | 67.3973 | oryctolagus_cuniculus | ENSOCUP00000045782 | 65.7895 | 58.8517 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.0822 | 71.5069 | oryctolagus_cuniculus | ENSOCUP00000031066 | 70.8955 | 64.9254 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.4521 | 71.7808 | equus_caballus | ENSECAP00000015938 | 79.235 | 71.5847 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 85.2055 | 77.5342 | equus_caballus | ENSECAP00000043671 | 77.3632 | 70.398 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 84.3836 | 77.5342 | equus_caballus | ENSECAP00000025114 | 84.6154 | 77.7473 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.0685 | 64.1096 | equus_caballus | ENSECAP00000014973 | 64.9289 | 55.4502 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 77.5342 | 69.863 | equus_caballus | ENSECAP00000042372 | 76.6938 | 69.1057 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.1918 | 76.7123 | equus_caballus | ENSECAP00000040463 | 55.7621 | 52.0446 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.8219 | 71.7808 | equus_caballus | ENSECAP00000047529 | 49.8311 | 44.2568 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 36.9863 | 33.4247 | equus_caballus | ENSECAP00000009540 | 68.8775 | 62.2449 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 69.863 | 61.3699 | canis_lupus_familiaris | ENSCAFP00845023758 | 68.3646 | 60.0536 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.4521 | 70.9589 | canis_lupus_familiaris | ENSCAFP00845023931 | 76.1155 | 67.979 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.9041 | 67.3973 | canis_lupus_familiaris | ENSCAFP00845013998 | 62.8821 | 53.7118 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 74.5205 | 64.9315 | canis_lupus_familiaris | ENSCAFP00845024169 | 54.0755 | 47.1173 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 29.589 | 24.6575 | canis_lupus_familiaris | ENSCAFP00845016737 | 59.6685 | 49.7238 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.1918 | 73.1507 | canis_lupus_familiaris | ENSCAFP00845013451 | 82.4176 | 73.3516 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 49.3151 | 43.5616 | panthera_pardus | ENSPPRP00000022214 | 78.6026 | 69.4323 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 46.0274 | 39.726 | panthera_pardus | ENSPPRP00000028186 | 79.2453 | 68.3962 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 73.1507 | 63.8356 | panthera_pardus | ENSPPRP00000008976 | 70.4485 | 61.4776 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 77.8082 | 68.7671 | panthera_pardus | ENSPPRP00000014457 | 76.7568 | 67.8378 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.7397 | 73.6986 | panthera_pardus | ENSPPRP00000001233 | 82.7397 | 73.6986 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 65.2055 | 57.5342 | panthera_pardus | ENSPPRP00000026957 | 67.2316 | 59.322 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 71.2329 | 61.3699 | panthera_pardus | ENSPPRP00000011135 | 75.1445 | 64.7399 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.4521 | 71.2329 | panthera_pardus | ENSPPRP00000001681 | 77.3333 | 69.3333 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 76.9863 | 67.3973 | panthera_pardus | ENSPPRP00000019311 | 76.776 | 67.2131 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 60.274 | 52.0548 | panthera_pardus | ENSPPRP00000015464 | 69.6203 | 60.1266 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 61.6438 | 55.8904 | panthera_pardus | ENSPPRP00000000843 | 82.1168 | 74.4526 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 62.1918 | 56.7123 | panthera_pardus | ENSPPRP00000011377 | 52.7907 | 48.1395 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 51.5069 | 45.2055 | panthera_pardus | ENSPPRP00000002320 | 78.0083 | 68.4647 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.3288 | 61.0959 | marmota_marmota_marmota | ENSMMMP00000026052 | 67.3469 | 56.8878 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 64.6575 | 55.3425 | marmota_marmota_marmota | ENSMMMP00000012382 | 68.0115 | 58.2133 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 87.1233 | 81.0959 | balaenoptera_musculus | ENSBMSP00010012509 | 87.6033 | 81.5427 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 84.3836 | 77.5342 | balaenoptera_musculus | ENSBMSP00010020379 | 80.8399 | 74.2782 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.274 | 71.5069 | felis_catus | ENSFCAP00000051531 | 58.7174 | 52.3046 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 64.1096 | 55.8904 | felis_catus | ENSFCAP00000018895 | 65.5462 | 57.1429 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.3562 | 69.3151 | felis_catus | ENSFCAP00000025615 | 80.112 | 70.8683 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 58.9041 | 49.863 | felis_catus | ENSFCAP00000015735 | 49.0868 | 41.5525 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.0548 | 62.7397 | felis_catus | ENSFCAP00000030752 | 74.0845 | 64.507 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 81.0959 | 71.5069 | felis_catus | ENSFCAP00000045112 | 72.9064 | 64.2857 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 35.8904 | 31.2329 | ursus_americanus | ENSUAMP00000036887 | 73.1844 | 63.6871 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 67.3973 | 63.8356 | ursus_americanus | ENSUAMP00000036029 | 68.5237 | 64.9025 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.8904 | 67.6712 | ursus_americanus | ENSUAMP00000005465 | 64.2691 | 57.3086 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.9041 | 68.4931 | ursus_americanus | ENSUAMP00000016352 | 80.4469 | 69.8324 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.5479 | 73.1507 | ursus_americanus | ENSUAMP00000006240 | 81.8941 | 74.3733 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.6164 | 72.0548 | ursus_americanus | ENSUAMP00000009584 | 82.1429 | 78.2738 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.8904 | 69.589 | ursus_americanus | ENSUAMP00000000976 | 57.5884 | 52.8067 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 43.0137 | 35.3425 | monodelphis_domestica | ENSMODP00000059064 | 67.6724 | 55.6034 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 37.5342 | 27.3973 | monodelphis_domestica | ENSMODP00000045298 | 54.8 | 40 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.6027 | 56.4384 | monodelphis_domestica | ENSMODP00000056841 | 60.0907 | 46.712 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 60.274 | 46.8493 | monodelphis_domestica | ENSMODP00000058105 | 60.7735 | 47.2376 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 65.7534 | 50.9589 | monodelphis_domestica | ENSMODP00000046044 | 66.2983 | 51.3812 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 68.2192 | 55.6164 | monodelphis_domestica | ENSMODP00000058836 | 64.0103 | 52.1851 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 67.3973 | 53.4247 | monodelphis_domestica | ENSMODP00000057213 | 65.4255 | 51.8617 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 62.4658 | 47.1233 | monodelphis_domestica | ENSMODP00000048504 | 55.0725 | 41.5459 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 46.5753 | 38.6301 | monodelphis_domestica | ENSMODP00000054745 | 72.3404 | 60 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 64.9315 | 53.1507 | monodelphis_domestica | ENSMODP00000018272 | 66.7606 | 54.6479 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 69.3151 | 53.4247 | monodelphis_domestica | ENSMODP00000059633 | 67.2872 | 51.8617 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 49.589 | 33.1507 | monodelphis_domestica | ENSMODP00000056096 | 48.1383 | 32.1809 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 62.7397 | 46.8493 | monodelphis_domestica | ENSMODP00000053892 | 60.105 | 44.8819 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 50.9589 | 46.0274 | bison_bison_bison | ENSBBBP00000013730 | 80.5195 | 72.7273 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.4521 | 70.411 | bison_bison_bison | ENSBBBP00000007802 | 60.0414 | 53.2091 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 35.3425 | 30.137 | bison_bison_bison | ENSBBBP00000018090 | 59.7222 | 50.9259 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.3288 | 62.1918 | bison_bison_bison | ENSBBBP00000018093 | 71.7391 | 61.6848 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 32.0548 | 25.4795 | bison_bison_bison | ENSBBBP00000005572 | 72.2222 | 57.4074 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 29.863 | 25.2055 | bison_bison_bison | ENSBBBP00000001702 | 59.2391 | 50 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.4658 | 75.0685 | bison_bison_bison | ENSBBBP00000010329 | 82.9201 | 75.4821 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 37.2603 | 30.411 | bison_bison_bison | ENSBBBP00000010299 | 63.2558 | 51.6279 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.726 | 72.8767 | bison_bison_bison | ENSBBBP00000001740 | 81.0585 | 74.0947 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 85.7534 | 79.1781 | monodon_monoceros | ENSMMNP00015016573 | 85.5191 | 78.9617 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 85.7534 | 78.3562 | monodon_monoceros | ENSMMNP00015015047 | 86.4641 | 79.0055 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 65.7534 | 56.7123 | sus_scrofa | ENSSSCP00000065664 | 53.6913 | 46.3087 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 68.4931 | 57.5342 | sus_scrofa | ENSSSCP00000001485 | 62.3441 | 52.3691 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.3288 | 63.5616 | sus_scrofa | ENSSSCP00000079792 | 77.4194 | 68.0352 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.4658 | 72.8767 | sus_scrofa | ENSSSCP00000044154 | 69.6759 | 61.5741 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 77.2603 | 67.1233 | sus_scrofa | ENSSSCP00000042783 | 76.4228 | 66.3957 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.6301 | 65.2055 | sus_scrofa | ENSSSCP00000049757 | 66.129 | 54.8387 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 69.589 | 63.0137 | sus_scrofa | ENSSSCP00000075103 | 68.2796 | 61.828 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 64.1096 | 54.2466 | sus_scrofa | ENSSSCP00000080524 | 66.4773 | 56.25 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 76.7123 | 66.5753 | ovis_aries_rambouillet | ENSOARP00020030013 | 74.8663 | 64.9733 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 77.2603 | 67.9452 | ovis_aries_rambouillet | ENSOARP00020024401 | 75.2 | 66.1333 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 76.4384 | 66.5753 | ovis_aries_rambouillet | ENSOARP00020021233 | 75.2022 | 65.4986 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.6164 | 66.8493 | ovis_aries_rambouillet | ENSOARP00020022751 | 72.2513 | 63.8743 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.726 | 70.6849 | ovis_aries_rambouillet | ENSOARP00020030225 | 75.7812 | 67.1875 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 76.1644 | 68.2192 | ovis_aries_rambouillet | ENSOARP00020024292 | 69.8492 | 62.5628 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 52.8767 | 45.4795 | ovis_aries_rambouillet | ENSOARP00020021302 | 70.1818 | 60.3636 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 61.6438 | 52.0548 | ovis_aries_rambouillet | ENSOARP00020015479 | 63.0252 | 53.2213 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.4658 | 75.6164 | ovis_aries_rambouillet | ENSOARP00020017643 | 64.1791 | 58.8486 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.8767 | 65.7534 | ovis_aries_rambouillet | ENSOARP00020024192 | 78.6982 | 71.0059 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.0822 | 67.9452 | ovis_aries_rambouillet | ENSOARP00020016303 | 73.6434 | 64.0827 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 71.5069 | 60.5479 | ovis_aries_rambouillet | ENSOARP00020021102 | 71.5069 | 60.5479 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 39.4521 | 32.8767 | ochotona_princeps | ENSOPRP00000004888 | 72.3618 | 60.3015 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 56.1644 | 49.863 | ochotona_princeps | ENSOPRP00000004731 | 82 | 72.8 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 42.4658 | 36.4384 | ochotona_princeps | ENSOPRP00000004215 | 75.2427 | 64.5631 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.8219 | 71.5069 | ochotona_princeps | ENSOPRP00000001469 | 81.9444 | 72.5 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 81.3699 | 70.137 | ochotona_princeps | ENSOPRP00000016362 | 83.427 | 71.9101 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 53.1507 | 46.3014 | ochotona_princeps | ENSOPRP00000011210 | 81.8565 | 71.308 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 69.589 | 60.5479 | ochotona_princeps | ENSOPRP00000002464 | 79.6238 | 69.279 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 23.5616 | 20.8219 | ochotona_princeps | ENSOPRP00000015120 | 64.1791 | 56.7164 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 58.3562 | 51.7808 | ochotona_princeps | ENSOPRP00000004209 | 59.0028 | 52.3546 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.6301 | 68.2192 | ochotona_princeps | ENSOPRP00000003094 | 79.0634 | 68.595 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 31.2329 | 28.2192 | ochotona_princeps | ENSOPRP00000011649 | 89.7638 | 81.1024 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 76.9863 | 67.1233 | ochotona_princeps | ENSOPRP00000015100 | 77.1978 | 67.3077 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 52.8767 | 45.2055 | ochotona_princeps | ENSOPRP00000015109 | 61.6613 | 52.7157 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 47.3973 | 42.7397 | ochotona_princeps | ENSOPRP00000015441 | 84.3902 | 76.0976 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 74.2466 | 63.8356 | ochotona_princeps | ENSOPRP00000010840 | 74.2466 | 63.8356 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 84.3836 | 76.4384 | delphinapterus_leucas | ENSDLEP00000026694 | 85.5556 | 77.5 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.9041 | 70.411 | physeter_catodon | ENSPCTP00005017546 | 78.9041 | 70.411 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 71.2329 | 61.6438 | dipodomys_ordii | ENSDORP00000003059 | 75.3623 | 65.2174 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 67.6712 | 57.8082 | dipodomys_ordii | ENSDORP00000006549 | 76.7081 | 65.528 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 50.6849 | 46.0274 | mustela_putorius_furo | ENSMPUP00000002067 | 69.5489 | 63.1579 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 68.7671 | 57.8082 | mustela_putorius_furo | ENSMPUP00000013884 | 70.3081 | 59.1036 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 73.1507 | 69.589 | aotus_nancymaae | ENSANAP00000006899 | 90.5085 | 86.1017 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 85.7534 | 78.9041 | aotus_nancymaae | ENSANAP00000007558 | 77.8607 | 71.6418 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.4658 | 74.5205 | aotus_nancymaae | ENSANAP00000009575 | 70 | 63.2558 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 84.1096 | 77.5342 | aotus_nancymaae | ENSANAP00000018586 | 83.6512 | 77.1117 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 85.4795 | 81.9178 | saimiri_boliviensis_boliviensis | ENSSBOP00000014539 | 77.037 | 73.8272 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 38.3562 | 34.5205 | vicugna_pacos | ENSVPAP00000006473 | 66.0377 | 59.434 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 41.0959 | 35.3425 | vicugna_pacos | ENSVPAP00000007229 | 76.5306 | 65.8163 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 71.7808 | 64.9315 | tupaia_belangeri | ENSTBEP00000006131 | 85.342 | 77.1987 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 39.726 | 36.7123 | tupaia_belangeri | ENSTBEP00000003189 | 70.7317 | 65.3659 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 52.8767 | 48.2192 | tupaia_belangeri | ENSTBEP00000007191 | 85.7778 | 78.2222 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 34.7945 | 32.0548 | tupaia_belangeri | ENSTBEP00000014087 | 68.2796 | 62.9032 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 35.0685 | 32.3288 | tupaia_belangeri | ENSTBEP00000002527 | 63.0542 | 58.1281 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 53.6986 | 49.3151 | tupaia_belangeri | ENSTBEP00000014199 | 54.1436 | 49.7238 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 38.9041 | 35.6164 | tupaia_belangeri | ENSTBEP00000009684 | 69.2683 | 63.4146 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 34.5205 | 31.5068 | tupaia_belangeri | ENSTBEP00000008033 | 67.7419 | 61.828 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 37.5342 | 34.7945 | tupaia_belangeri | ENSTBEP00000011531 | 65.5502 | 60.7655 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 34.2466 | 32.0548 | tupaia_belangeri | ENSTBEP00000006715 | 67.5676 | 63.2432 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 35.6164 | 32.8767 | tupaia_belangeri | ENSTBEP00000007977 | 63.7255 | 58.8235 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 30.137 | 27.1233 | tupaia_belangeri | ENSTBEP00000001222 | 88 | 79.2 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 19.1781 | 16.4384 | tupaia_belangeri | ENSTBEP00000001987 | 33.6538 | 28.8462 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 56.1644 | 52.6027 | phocoena_sinus | ENSPSNP00000022900 | 84.7107 | 79.3388 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.1918 | 75.6164 | phocoena_sinus | ENSPSNP00000014177 | 77.7202 | 71.5026 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 37.2603 | 33.4247 | camelus_dromedarius | ENSCDRP00005024197 | 62.6728 | 56.2212 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 69.589 | 62.7397 | camelus_dromedarius | ENSCDRP00005021722 | 68.0965 | 61.3941 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 70.6849 | 64.6575 | camelus_dromedarius | ENSCDRP00005016911 | 82.4281 | 75.3994 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 61.3699 | 55.3425 | camelus_dromedarius | ENSCDRP00005016654 | 62.7451 | 56.5826 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.1918 | 74.5205 | camelus_dromedarius | ENSCDRP00005029070 | 81.7439 | 74.1144 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 73.9726 | 66.3014 | carlito_syrichta | ENSTSYP00000015499 | 77.3639 | 69.341 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 53.6986 | 48.7671 | carlito_syrichta | ENSTSYP00000005254 | 83.4043 | 75.7447 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.6164 | 67.1233 | carlito_syrichta | ENSTSYP00000025861 | 72.2513 | 64.1361 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.6301 | 67.9452 | panthera_tigris_altaica | ENSPTIP00000005576 | 77.3585 | 66.8464 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 73.9726 | 65.4795 | panthera_tigris_altaica | ENSPTIP00000020192 | 77.5862 | 68.6782 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.1918 | 73.9726 | panthera_tigris_altaica | ENSPTIP00000003081 | 82.8729 | 74.5856 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 69.863 | 62.1918 | panthera_tigris_altaica | ENSPTIP00000003142 | 72.8571 | 64.8571 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 32.0548 | 27.9452 | panthera_tigris_altaica | ENSPTIP00000003157 | 76.4706 | 66.6667 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.726 | 70.411 | panthera_tigris_altaica | ENSPTIP00000003396 | 75.9791 | 67.1018 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 70.411 | 62.1918 | panthera_tigris_altaica | ENSPTIP00000009178 | 62.2276 | 54.9637 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 66.3014 | 56.9863 | peromyscus_maniculatus_bairdii | ENSPEMP00000031428 | 69.1429 | 59.4286 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.274 | 71.5069 | bos_mutus | ENSBMUP00000017841 | 61.0417 | 54.375 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 76.4384 | 67.1233 | bos_mutus | ENSBMUP00000030164 | 62.6966 | 55.0562 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 46.5753 | 41.6438 | bos_mutus | ENSBMUP00000002961 | 70.8333 | 63.3333 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 52.6027 | 45.2055 | bos_mutus | ENSBMUP00000022247 | 70.5882 | 60.6618 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 70.9589 | 63.5616 | bos_mutus | ENSBMUP00000009484 | 68.7003 | 61.5385 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 49.0411 | 40.274 | bos_mutus | ENSBMUP00000017874 | 71.8876 | 59.0361 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 31.2329 | 26.8493 | bos_mutus | ENSBMUP00000030078 | 67.0588 | 57.6471 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 33.4247 | 27.9452 | bos_mutus | ENSBMUP00000030058 | 48.8 | 40.8 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 41.0959 | 34.7945 | bos_mutus | ENSBMUP00000035923 | 43.9883 | 37.2434 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.5479 | 74.2466 | myotis_lucifugus | ENSMLUP00000010929 | 80.3279 | 74.0437 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 76.4384 | 67.9452 | myotis_lucifugus | ENSMLUP00000019084 | 79.2614 | 70.4545 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.8219 | 72.3288 | myotis_lucifugus | ENSMLUP00000000544 | 80.8219 | 72.3288 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.274 | 72.8767 | myotis_lucifugus | ENSMLUP00000022087 | 80.0546 | 72.6776 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 51.5069 | 46.5753 | myotis_lucifugus | ENSMLUP00000010932 | 79.661 | 72.0339 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.6164 | 67.3973 | myotis_lucifugus | ENSMLUP00000021164 | 79.5389 | 70.8934 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 81.3699 | 74.2466 | myotis_lucifugus | ENSMLUP00000014385 | 77.1429 | 70.3896 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.3562 | 71.5069 | myotis_lucifugus | ENSMLUP00000021135 | 80.112 | 73.1092 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.6164 | 66.8493 | myotis_lucifugus | ENSMLUP00000002042 | 78.187 | 69.1218 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 81.9178 | 73.4247 | myotis_lucifugus | ENSMLUP00000018286 | 81.694 | 73.224 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.1918 | 74.2466 | myotis_lucifugus | ENSMLUP00000020003 | 82.4176 | 74.4505 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 54.7945 | 49.589 | myotis_lucifugus | ENSMLUP00000018369 | 82.6446 | 74.7934 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.5479 | 71.7808 | myotis_lucifugus | ENSMLUP00000008271 | 79.6748 | 71.0027 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 81.3699 | 72.8767 | myotis_lucifugus | ENSMLUP00000022041 | 81.8182 | 73.2782 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.8219 | 74.7945 | myotis_lucifugus | ENSMLUP00000022295 | 80.8219 | 74.7945 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 65.2055 | 60.274 | myotis_lucifugus | ENSMLUP00000000091 | 80.4054 | 74.3243 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.5479 | 73.1507 | myotis_lucifugus | ENSMLUP00000009864 | 80.3279 | 72.9508 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.7397 | 74.5205 | myotis_lucifugus | ENSMLUP00000012602 | 82.7397 | 74.5205 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 61.6438 | 54.2466 | myotis_lucifugus | ENSMLUP00000018781 | 69.0184 | 60.7362 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.3562 | 70.6849 | myotis_lucifugus | ENSMLUP00000005486 | 78.3562 | 70.6849 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 76.7123 | 69.0411 | myotis_lucifugus | ENSMLUP00000021461 | 78.8732 | 70.9859 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.8219 | 72.8767 | myotis_lucifugus | ENSMLUP00000000039 | 79.7297 | 71.8919 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 41.3699 | 34.5205 | myotis_lucifugus | ENSMLUP00000017999 | 79.0576 | 65.9686 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.9041 | 70.6849 | myotis_lucifugus | ENSMLUP00000020562 | 72.7273 | 65.1515 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.9041 | 70.137 | myotis_lucifugus | ENSMLUP00000016745 | 79.558 | 70.7182 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 63.8356 | 57.2603 | myotis_lucifugus | ENSMLUP00000021310 | 71.6923 | 64.3077 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.726 | 72.3288 | myotis_lucifugus | ENSMLUP00000006112 | 79.5082 | 72.1311 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.274 | 70.6849 | myotis_lucifugus | ENSMLUP00000022414 | 81.3889 | 71.6667 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.8219 | 72.3288 | myotis_lucifugus | ENSMLUP00000013245 | 80.6011 | 72.1311 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 54.2466 | 48.4931 | myotis_lucifugus | ENSMLUP00000021330 | 81.8182 | 73.1405 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.274 | 71.7808 | myotis_lucifugus | ENSMLUP00000002414 | 80.0546 | 71.5847 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 69.3151 | 59.1781 | myotis_lucifugus | ENSMLUP00000020659 | 75.976 | 64.8649 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 73.6986 | 66.0274 | myotis_lucifugus | ENSMLUP00000017958 | 79.822 | 71.5134 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80 | 70.9589 | myotis_lucifugus | ENSMLUP00000022602 | 79.3478 | 70.3804 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.6301 | 69.863 | myotis_lucifugus | ENSMLUP00000022801 | 71.5711 | 63.591 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.6027 | 62.4658 | myotis_lucifugus | ENSMLUP00000010307 | 74.4382 | 64.0449 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.5479 | 72.0548 | myotis_lucifugus | ENSMLUP00000016430 | 80.109 | 71.6621 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 81.3699 | 74.5205 | myotis_lucifugus | ENSMLUP00000021115 | 81.3699 | 74.5205 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 63.0137 | 57.2603 | myotis_lucifugus | ENSMLUP00000000662 | 77.9661 | 70.8475 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.726 | 70.6849 | myotis_lucifugus | ENSMLUP00000008096 | 79.5082 | 70.4918 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 53.1507 | 49.589 | myotis_lucifugus | ENSMLUP00000008009 | 80.4979 | 75.1037 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.4658 | 74.2466 | myotis_lucifugus | ENSMLUP00000017409 | 82.2404 | 74.0437 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 37.8082 | 34.2466 | myotis_lucifugus | ENSMLUP00000012709 | 87.3418 | 79.1139 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.8219 | 74.2466 | myotis_lucifugus | ENSMLUP00000015384 | 79.3011 | 72.8495 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 62.4658 | 54.2466 | neovison_vison | ENSNVIP00000021341 | 63.6871 | 55.3073 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 56.4384 | 48.7671 | neovison_vison | ENSNVIP00000017461 | 67.9868 | 58.7459 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 68.4931 | 62.7397 | neovison_vison | ENSNVIP00000023634 | 62.0347 | 56.8238 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 83.2877 | 76.1644 | neovison_vison | ENSNVIP00000021545 | 84.2105 | 77.0083 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.8767 | 62.4658 | neovison_vison | ENSNVIP00000025985 | 70.9333 | 60.8 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 72.0548 | 64.1096 | bos_taurus | ENSBTAP00000071237 | 59.9089 | 53.303 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 81.0959 | 72.8767 | bos_taurus | ENSBTAP00000070120 | 75.5102 | 67.8571 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 83.8356 | 76.7123 | bos_taurus | ENSBTAP00000035745 | 70.0229 | 64.0732 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 66.3014 | 56.7123 | bos_taurus | ENSBTAP00000066745 | 62.0513 | 53.0769 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 77.2603 | 68.7671 | bos_taurus | ENSBTAP00000016194 | 78.771 | 70.1117 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 81.0959 | 72.0548 | bos_taurus | ENSBTAP00000059934 | 81.5427 | 72.4518 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 64.9315 | 58.9041 | bos_taurus | ENSBTAP00000074509 | 77.451 | 70.2614 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.0685 | 67.6712 | bos_taurus | ENSBTAP00000021451 | 74.6594 | 67.3025 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 65.2055 | 58.6301 | canis_lupus_dingo | ENSCAFP00020026118 | 66.4804 | 59.7765 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 73.4247 | 64.3836 | canis_lupus_dingo | ENSCAFP00020025619 | 64.891 | 56.9007 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 31.7808 | 26.8493 | canis_lupus_dingo | ENSCAFP00020016937 | 63.388 | 53.5519 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 73.4247 | 64.1096 | canis_lupus_dingo | ENSCAFP00020033194 | 61.7512 | 53.917 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80 | 70.137 | canis_lupus_dingo | ENSCAFP00020025627 | 74.4898 | 65.3061 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 75.0685 | 63.2877 | canis_lupus_dingo | ENSCAFP00020026073 | 72.2955 | 60.9499 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 70.9589 | 62.1918 | procavia_capensis | ENSPCAP00000000453 | 74 | 64.8571 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 67.9452 | 60.8219 | procavia_capensis | ENSPCAP00000015575 | 80 | 71.6129 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 69.0411 | 61.9178 | procavia_capensis | ENSPCAP00000004010 | 70.5882 | 63.3053 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 33.6986 | 31.2329 | procavia_capensis | ENSPCAP00000000492 | 62.1212 | 57.5758 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 35.3425 | 31.7808 | procavia_capensis | ENSPCAP00000007166 | 61.1374 | 54.9763 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 66.8493 | 58.9041 | procavia_capensis | ENSPCAP00000002139 | 78.9644 | 69.5793 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 37.2603 | 33.9726 | procavia_capensis | ENSPCAP00000012243 | 83.4356 | 76.0736 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 34.5205 | 31.5068 | procavia_capensis | ENSPCAP00000002911 | 88.7324 | 80.9859 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 35.8904 | 31.2329 | bos_grunniens | ENSBGRP00000020046 | 78.9157 | 68.6747 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 78.9041 | 71.7808 | bos_grunniens | ENSBGRP00000034456 | 67.4473 | 61.3583 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 56.1644 | 49.589 | bos_grunniens | ENSBGRP00000018860 | 65.7051 | 58.0128 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 66.5753 | 58.6301 | bos_grunniens | ENSBGRP00000015990 | 65.1475 | 57.3727 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 76.4384 | 68.7671 | bos_grunniens | ENSBGRP00000019882 | 83.2836 | 74.9254 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 76.1644 | 69.589 | cervus_hanglu_yarkandensis | ENSCHYP00000013724 | 83.4835 | 76.2763 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 70.9589 | 60.274 | cervus_hanglu_yarkandensis | ENSCHYP00000032846 | 62.4096 | 53.012 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 81.9178 | 72.3288 | cervus_hanglu_yarkandensis | ENSCHYP00000002805 | 65.859 | 58.1498 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 52.6027 | 43.2877 | cervus_hanglu_yarkandensis | ENSCHYP00000024480 | 47.5248 | 39.1089 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 32.0548 | 26.8493 | cervus_hanglu_yarkandensis | ENSCHYP00000015857 | 75.4839 | 63.2258 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 73.6986 | 67.6712 | cervus_hanglu_yarkandensis | ENSCHYP00000016863 | 76.204 | 69.9717 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80 | 70.9589 | cervus_hanglu_yarkandensis | ENSCHYP00000000469 | 71.7445 | 63.6364 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 71.2329 | 63.8356 | cervus_hanglu_yarkandensis | ENSCHYP00000002980 | 80 | 71.6923 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 71.7808 | 62.1918 | cervus_hanglu_yarkandensis | ENSCHYP00000028036 | 72.9805 | 63.2312 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 67.3973 | 57.2603 | cervus_hanglu_yarkandensis | ENSCHYP00000027891 | 72.3529 | 61.4706 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 73.6986 | 63.8356 | cervus_hanglu_yarkandensis | ENSCHYP00000029191 | 60.179 | 52.1253 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 55.0685 | 48.7671 | catagonus_wagneri | ENSCWAP00000007352 | 73.6264 | 65.2015 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 67.3973 | 56.9863 | catagonus_wagneri | ENSCWAP00000007309 | 75.6923 | 64 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 70.137 | 61.0959 | ictidomys_tridecemlineatus | ENSSTOP00000017429 | 74.8538 | 65.2047 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 57.5342 | 51.5069 | otolemur_garnettii | ENSOGAP00000015293 | 83.3333 | 74.6032 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.274 | 70.137 | otolemur_garnettii | ENSOGAP00000018061 | 80.9392 | 70.7182 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.274 | 70.6849 | otolemur_garnettii | ENSOGAP00000022324 | 83.4758 | 73.5043 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 83.5616 | 77.8082 | otolemur_garnettii | ENSOGAP00000014883 | 85.1955 | 79.3296 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 85.2055 | 77.5342 | otolemur_garnettii | ENSOGAP00000020581 | 85.6749 | 77.9614 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 86.0274 | 77.5342 | otolemur_garnettii | ENSOGAP00000016376 | 86.2637 | 77.7473 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.726 | 72.8767 | otolemur_garnettii | ENSOGAP00000022187 | 82.6705 | 75.5682 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 86.8493 | 80 | otolemur_garnettii | ENSOGAP00000006055 | 87.3278 | 80.4408 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 83.0137 | 76.4384 | otolemur_garnettii | ENSOGAP00000003699 | 83.7017 | 77.0718 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 81.3699 | 74.5205 | otolemur_garnettii | ENSOGAP00000017874 | 80.7065 | 73.913 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 82.1918 | 74.7945 | otolemur_garnettii | ENSOGAP00000020548 | 83.3333 | 75.8333 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.8219 | 73.1507 | otolemur_garnettii | ENSOGAP00000010481 | 87.0207 | 78.7611 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 86.8493 | 82.1918 | cebus_imitator | ENSCCAP00000040455 | 70.9172 | 67.1141 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 60.8219 | 51.5069 | urocitellus_parryii | ENSUPAP00010001881 | 57.6623 | 48.8312 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 79.1781 | 70.411 | moschus_moschiferus | ENSMMSP00000007999 | 78.1081 | 69.4595 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 80.5479 | 70.6849 | moschus_moschiferus | ENSMMSP00000024009 | 76.5625 | 67.1875 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 28.4932 | 17.2603 | callorhinchus_milii | ENSCMIP00000005770 | 55.615 | 33.6898 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 49.0411 | 32.6027 | callorhinchus_milii | ENSCMIP00000012667 | 43.3414 | 28.8136 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 27.3973 | 17.2603 | latimeria_chalumnae | ENSLACP00000001377 | 44.2478 | 27.8761 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 23.0137 | 13.4247 | latimeria_chalumnae | ENSLACP00000006880 | 39.6226 | 23.1132 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 36.1644 | 21.0959 | lepisosteus_oculatus | ENSLOCP00000000811 | 49.0706 | 28.6245 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 22.1918 | 12.6027 | danio_rerio | ENSDARP00000128245 | 37.156 | 21.1009 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 24.1096 | 14.2466 | danio_rerio | ENSDARP00000157157 | 34.6457 | 20.4724 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 31.7808 | 20.274 | carassius_auratus | ENSCARP00000003279 | 41.8773 | 26.7148 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 26.5753 | 16.1644 | laticauda_laticaudata | ENSLLTP00000013476 | 47.549 | 28.9216 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 22.1918 | 16.1644 | notechis_scutatus | ENSNSUP00000002776 | 60.4478 | 44.0299 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 18.3562 | 13.1507 | notechis_scutatus | ENSNSUP00000005301 | 62.037 | 44.4444 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 17.8082 | 12.0548 | notechis_scutatus | ENSNSUP00000000959 | 56.0345 | 37.931 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 18.6301 | 13.6986 | notechis_scutatus | ENSNSUP00000002214 | 50.3704 | 37.037 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 16.1644 | 11.7808 | notechis_scutatus | ENSNSUP00000000612 | 67.0455 | 48.8636 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 53.4247 | 37.2603 | gallus_gallus | ENSGALP00010005066 | 44.6224 | 31.1213 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 52.0548 | 36.4384 | gallus_gallus | ENSGALP00010003418 | 49.8688 | 34.9081 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 48.4931 | 33.1507 | gallus_gallus | ENSGALP00010003388 | 41.3551 | 28.271 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 53.6986 | 37.2603 | gallus_gallus | ENSGALP00010005104 | 51.715 | 35.8839 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 52.3288 | 37.5342 | gallus_gallus | ENSGALP00010005075 | 48.7245 | 34.949 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 53.6986 | 37.8082 | gallus_gallus | ENSGALP00010005186 | 56.9767 | 40.1163 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 54.2466 | 37.5342 | gallus_gallus | ENSGALP00010005093 | 40.3259 | 27.9022 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 38.6301 | 27.9452 | gallus_gallus | ENSGALP00010000192 | 46.0784 | 33.3333 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 52.0548 | 36.4384 | gallus_gallus | ENSGALP00010003108 | 54.1311 | 37.8917 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 54.2466 | 37.5342 | gallus_gallus | ENSGALP00010003454 | 40.3259 | 27.9022 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 51.7808 | 38.0822 | gallus_gallus | ENSGALP00010003530 | 42.4719 | 31.236 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 54.5205 | 39.1781 | gallus_gallus | ENSGALP00010005134 | 48.8943 | 35.1351 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 54.7945 | 38.9041 | gallus_gallus | ENSGALP00010005027 | 53.4759 | 37.9679 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 23.5616 | 15.8904 | gallus_gallus | ENSGALP00010000194 | 39.8148 | 26.8519 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 53.4247 | 37.5342 | gallus_gallus | ENSGALP00010005042 | 51.8617 | 36.4362 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 35.3425 | 24.9315 | gallus_gallus | ENSGALP00010003089 | 39.2097 | 27.6596 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 56.1644 | 38.3562 | gallus_gallus | ENSGALP00010004373 | 56.9444 | 38.8889 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 26.3014 | 17.5342 | gallus_gallus | ENSGALP00010003079 | 53.9326 | 35.9551 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 53.4247 | 38.3562 | gallus_gallus | ENSGALP00010003554 | 53.8674 | 38.674 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 47.9452 | 32.8767 | gallus_gallus | ENSGALP00010002851 | 54.8589 | 37.6176 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 53.4247 | 37.2603 | gallus_gallus | ENSGALP00010003022 | 42.1166 | 29.3736 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 30.6849 | 20.274 | parus_major | ENSPMJP00000004421 | 50.2242 | 33.1839 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 15.6164 | 10.6849 | anolis_carolinensis | ENSACAP00000018273 | 44.186 | 30.2326 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 17.5342 | 12.8767 | naja_naja | ENSNNAP00000017655 | 52.0325 | 38.2114 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 17.5342 | 12.6027 | podarcis_muralis | ENSPMRP00000001000 | 52.8926 | 38.0165 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 16.1644 | 9.86301 | salvator_merianae | ENSSMRP00000004481 | 36.4198 | 22.2222 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 17.2603 | 11.2329 | salvator_merianae | ENSSMRP00000004955 | 50 | 32.5397 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 17.5342 | 10.411 | oryzias_sinensis | ENSOSIP00000025859 | 49.6124 | 29.4574 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 15.6164 | 10.137 | oryzias_sinensis | ENSOSIP00000002372 | 50.8929 | 33.0357 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 35.3425 | 23.2877 | pelodiscus_sinensis | ENSPSIP00000017578 | 53.75 | 35.4167 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 15.6164 | 8.76712 | gopherus_evgoodei | ENSGEVP00005020513 | 31.4917 | 17.6796 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 49.589 | 35.6164 | anser_brachyrhynchus | ENSABRP00000001822 | 54.0299 | 38.806 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 43.8356 | 29.589 | anser_brachyrhynchus | ENSABRP00000001840 | 33.6842 | 22.7368 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 42.4658 | 27.9452 | anser_brachyrhynchus | ENSABRP00000027027 | 42.4658 | 27.9452 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 21.0959 | 13.9726 | gasterosteus_aculeatus | ENSGACP00000002564 | 52.381 | 34.6939 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 52.6027 | 37.5342 | strigops_habroptila | ENSSHBP00005001035 | 54.0845 | 38.5915 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 52.6027 | 37.2603 | strigops_habroptila | ENSSHBP00005008291 | 49.7409 | 35.2332 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 25.7534 | 16.7123 | stegastes_partitus | ENSSPAP00000027794 | 37.751 | 24.498 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 21.6438 | 12.3288 | oncorhynchus_tshawytscha | ENSOTSP00005083033 | 41.5789 | 23.6842 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 53.4247 | 38.9041 | aquila_chrysaetos_chrysaetos | ENSACCP00020024696 | 47.561 | 34.6341 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 31.7808 | 23.0137 | aquila_chrysaetos_chrysaetos | ENSACCP00020024706 | 48.1328 | 34.8548 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 12.3288 | 7.94521 | ficedula_albicollis | ENSFALP00000031472 | 34.3511 | 22.1374 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 49.863 | 37.2603 | coturnix_japonica | ENSCJPP00005019348 | 50.9804 | 38.0952 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 44.3836 | 31.5068 | coturnix_japonica | ENSCJPP00005019355 | 52.2581 | 37.0968 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 41.0959 | 25.7534 | coturnix_japonica | ENSCJPP00005019370 | 45.4545 | 28.4848 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 24.1096 | 17.2603 | tetraodon_nigroviridis | ENSTNIP00000006764 | 42.9268 | 30.7317 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 23.2877 | 16.1644 | tetraodon_nigroviridis | ENSTNIP00000004370 | 43.8144 | 30.4124 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 13.1507 | 9.0411 | paramormyrops_kingsleyae | ENSPKIP00000000612 | 41.7391 | 28.6957 |
| ENSG00000206503 | homo_sapiens | ENSP00000366005 | 44.3836 | 29.0411 | leptobrachium_leishanense | ENSLLEP00000039939 | 45.7627 | 29.9435 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000139 | located_in Golgi membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0001913 | involved_in T cell mediated cytotoxicity | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0001916 | involved_in positive regulation of T cell mediated cytotoxicity | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0002419 | involved_in T cell mediated cytotoxicity directed against tumor cell target | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0002476 | involved_in antigen processing and presentation of endogenous peptide antigen via MHC class Ib | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0002485 | involved_in antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0002486 | involved_in antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0002726 | involved_in positive regulation of T cell cytokine production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0005102 | enables signaling receptor binding | 2 | IBA,IPI | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005615 | is_active_in extracellular space | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0005783 | located_in endoplasmic reticulum | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005789 | located_in endoplasmic reticulum membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005794 | located_in Golgi apparatus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005797 | located_in Golgi medial cisterna | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005886 | located_in plasma membrane | 3 | IDA,NAS,TAS | Homo_sapiens(9606) | Component |
| GO:0006955 | involved_in immune response | 3 | IBA,IMP,NAS | Homo_sapiens(9606) | Process |
| GO:0009897 | is_active_in external side of plasma membrane | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0009986 | located_in cell surface | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0012507 | located_in ER to Golgi transport vesicle membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0016020 | located_in membrane | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0016045 | involved_in detection of bacterium | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0019731 | involved_in antibacterial humoral response | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0019885 | involved_in antigen processing and presentation of endogenous peptide antigen via MHC class I | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0030670 | located_in phagocytic vesicle membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0030881 | enables beta-2-microglobulin binding | 3 | IC,IDA,IMP | Homo_sapiens(9606) | Function |
| GO:0031901 | located_in early endosome membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0032729 | involved_in positive regulation of type II interferon production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0036037 | involved_in CD8-positive, alpha-beta T cell activation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0042270 | NOT involved_in protection from natural killer cell mediated cytotoxicity | 2 | IDA | Homo_sapiens(9606) | Process |
| GO:0042590 | involved_in antigen processing and presentation of exogenous peptide antigen via MHC class I | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0042605 | enables peptide antigen binding | 3 | IBA,IDA,IMP | Homo_sapiens(9606) | Function |
| GO:0042608 | enables T cell receptor binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0042610 | enables CD8 receptor binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0042612 | part_of MHC class I protein complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0042824 | part_of MHC class I peptide loading complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0045087 | involved_in innate immune response | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0046977 | enables TAP binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0050830 | involved_in defense response to Gram-positive bacterium | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0050852 | involved_in T cell receptor signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0055038 | located_in recycling endosome membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0062061 | enables TAP complex binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0070062 | located_in extracellular exosome | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0070971 | located_in endoplasmic reticulum exit site | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0098553 | located_in lumenal side of endoplasmic reticulum membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:2000566 | involved_in positive regulation of CD8-positive, alpha-beta T cell proliferation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:2000568 | involved_in positive regulation of memory T cell activation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:2001187 | involved_in positive regulation of CD8-positive, alpha-beta T cell activation | 1 | IDA | Homo_sapiens(9606) | Process |