RBP Type
Canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
E3 ligase
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000204610 Gene ID
89870 Accession
16284
Symbol
TRIM15
Alias
RNF93;ZNFB7;ZNF178
Full Name
tripartite motif containing 15
Status
Confidence
Length
9488 bases
Strand
Plus strand
Position
6 : 30163206 - 30172693
RNA binding domain
SPRY
Summary
The protein encoded by this gene is a member of the tripartite motif (TRIM) family. The TRIM motif includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. The protein localizes to the cytoplasm. Alternatively spliced transcript variants have been described, but their biological validity has not been determined. [provided by RefSeq, Jul 2008]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000484001
SPRY
PF00622 7.1e-11
1
ENSP00000365884
SPRY
PF00622 1.1e-10
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
12601 Psoriasis 1.675E-17 19680446 12919 Multiple Sclerosis 1.3E-5 19010793 12927 Multiple Sclerosis 1.7E-7 19010793 52144 Child Development Disorders, Pervasive 2.7000000E-005 - 52150 Child Development Disorders, Pervasive 2.4000000E-005 - 52163 Child Development Disorders, Pervasive 2.9000000E-005 - 52164 Child Development Disorders, Pervasive 3.3000000E-005 - 52176 Child Development Disorders, Pervasive 7.3600000E-006 - 52180 Child Development Disorders, Pervasive 2.9000000E-005 - 52195 Child Development Disorders, Pervasive 7.8500000E-006 - 52197 Child Development Disorders, Pervasive 3.3000000E-005 -
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
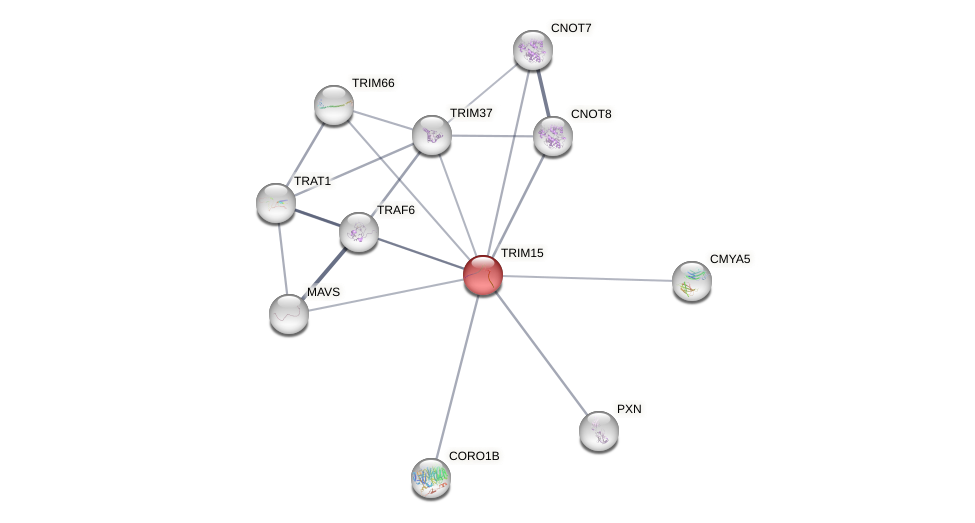
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0003713 enables transcription coactivator activity 1 IDA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 is_active_in nucleus 1 IDA Homo_sapiens(9606) Component GO:0005737 is_active_in cytoplasm 1 IBA Homo_sapiens(9606) Component GO:0005925 located_in focal adhesion 1 IEA Homo_sapiens(9606) Component GO:0007500 involved_in mesodermal cell fate determination 1 TAS Homo_sapiens(9606) Process GO:0008270 enables zinc ion binding 1 IEA Homo_sapiens(9606) Function GO:0016567 involved_in protein ubiquitination 1 IBA Homo_sapiens(9606) Process GO:0032481 involved_in positive regulation of type I interferon production 1 IMP Homo_sapiens(9606) Process GO:0044790 involved_in suppression of viral release by host 2 IDA,IMP Homo_sapiens(9606) Process GO:0045087 involved_in innate immune response 1 IMP Homo_sapiens(9606) Process GO:0045893 involved_in positive regulation of DNA-templated transcription 1 IEA Homo_sapiens(9606) Process GO:0051091 involved_in positive regulation of DNA-binding transcription factor activity 1 IMP Homo_sapiens(9606) Process GO:0051092 involved_in positive regulation of NF-kappaB transcription factor activity 2 IBA,IDA Homo_sapiens(9606) Process GO:0061630 enables ubiquitin protein ligase activity 1 IBA Homo_sapiens(9606) Function GO:0140313 enables molecular sequestering activity 1 IDA Homo_sapiens(9606) Function GO:1900246 involved_in positive regulation of RIG-I signaling pathway 1 IMP Homo_sapiens(9606) Process GO:1901223 involved_in negative regulation of NIK/NF-kappaB signaling 1 IDA Homo_sapiens(9606) Process GO:1901253 involved_in negative regulation of intracellular transport of viral material 1 IMP Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.