Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000197299 | Gene ID | 641 | Accession | 1058 |
| Symbol | BLM | Alias | BS;RECQ2;RECQL2;RECQL3;MGRISCE1 | Full Name | BLM RecQ like helicase |
| Status | Confidence | Length | 98821 bases | Strand | Plus strand |
| Position | 15 : 90717346 - 90816166 | RNA binding domain | BDHCT , DEAD , HRDC | ||
| Summary | The Bloom syndrome is an autosomal recessive disorder characterized by growth deficiency, microcephaly and immunodeficiency among others. It is caused by homozygous or compound heterozygous mutation in the gene encoding DNA helicase RecQ protein on chromosome 15q26. This Bloom-associated helicase unwinds a variety of DNA substrates including Holliday junction, and is involved in several pathways contributing to the maintenance of genome stability. Identification of pathogenic Bloom variants is required for heterozygote testing in at-risk families. [provided by RefSeq, May 2020] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000506682 | DEAD | PF00270 | 5.9e-19 | 1 |
| ENSP00000506682 | HRDC | PF00570 | 2.2e-11 | 2 |
| ENSP00000506682 | BDHCT | PF08072 | 3.1e-21 | 3 |
| ENSP00000497646 | DEAD | PF00270 | 6e-19 | 1 |
| ENSP00000497646 | HRDC | PF00570 | 2.2e-11 | 2 |
| ENSP00000497646 | BDHCT | PF08072 | 3.1e-21 | 3 |
| ENSP00000347232 | DEAD | PF00270 | 6e-19 | 1 |
| ENSP00000347232 | HRDC | PF00570 | 2.3e-11 | 2 |
| ENSP00000347232 | BDHCT | PF08072 | 3.3e-21 | 3 |
| ENSP00000506117 | DEAD | PF00270 | 6e-19 | 1 |
| ENSP00000506117 | HRDC | PF00570 | 2.3e-11 | 2 |
| ENSP00000506117 | BDHCT | PF08072 | 3.3e-21 | 3 |
| ENSP00000454158 | DEAD | PF00270 | 6.4e-19 | 1 |
| ENSP00000454158 | BDHCT | PF08072 | 2.8e-21 | 2 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 37503837 | BLM helicase protein negatively regulates stress granule formation through unwinding RNA G-quadruplex structures. | M Yehuda Danino | 2023-07-28 | Nucleic acids research |
| 36884289 | The nuclear isoforms of the Fragile X mental retardation RNA-binding protein associate with genomic DNA bridges. | N Ledoux | 2023-05-01 | Molecular biology of the cell |
| 36569948 | The BLM helicase is a new therapeutic target in multiple myeloma involved in replication stress survival and drug resistance. | Sara Ovejero | 2022-01-01 | Frontiers in immunology |
| 35440629 | A POLD3/BLM dependent pathway handles DSBs in transcribed chromatin upon excessive RNA:DNA hybrid accumulation. | S Cohen | 2022-04-19 | Nature communications |
| 36484368 | N6‑methyladenosine‑induced long non‑coding RNA PVT1 regulates the miR‑27b‑3p/BLM axis to promote prostate cancer progression. | Bin Chen | 2023-01-01 | International journal of oncology |
| 25245947 | RecQ-core of BLM unfolds telomeric G-quadruplex in the absence of ATP. | B Jagat Budhathoki | 2014-10-01 | Nucleic acids research |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| BLM-201 | ENST00000355112 | 5240 | 1417aa | ENSP00000347232 |
| BLM-213 | ENST00000681142 | 4270 | 1372aa | ENSP00000506682 |
| BLM-211 | ENST00000648453 | 4522 | 1386aa | ENSP00000497646 |
| BLM-208 | ENST00000560509 | 3966 | 1286aa | ENSP00000454158 |
| BLM-206 | ENST00000559724 | 4510 | 413aa | ENSP00000453359 |
| BLM-212 | ENST00000680772 | 5033 | 1417aa | ENSP00000506117 |
| BLM-204 | ENST00000559282 | 866 | No protein | - |
| BLM-202 | ENST00000558599 | 577 | No protein | - |
| BLM-205 | ENST00000559426 | 533 | No protein | - |
| BLM-207 | ENST00000560136 | 2023 | No protein | - |
| BLM-209 | ENST00000560559 | 2967 | No protein | - |
| BLM-203 | ENST00000558825 | 1683 | No protein | - |
| BLM-210 | ENST00000560821 | 1024 | No protein | - |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
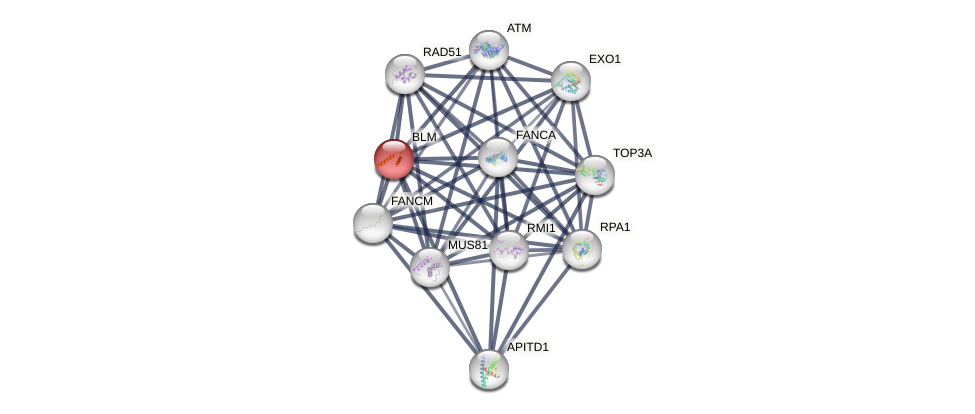
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000197299 | homo_sapiens | ENSP00000416739 | 54.2373 | 35.4391 | homo_sapiens | ENSP00000347232 | 24.8412 | 16.2315 |
| ENSG00000197299 | homo_sapiens | ENSP00000482313 | 30.2152 | 16.9702 | homo_sapiens | ENSP00000347232 | 25.7586 | 14.4672 |
| ENSG00000197299 | homo_sapiens | ENSP00000298139 | 32.3324 | 18.6453 | homo_sapiens | ENSP00000347232 | 32.6747 | 18.8426 |
| ENSG00000197299 | homo_sapiens | ENSP00000317636 | 36.7306 | 23.5116 | homo_sapiens | ENSP00000347232 | 25.6881 | 16.4432 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 97.3888 | 96.4008 | nomascus_leucogenys | ENSNLEP00000014738 | 98.6419 | 97.6412 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 99.4354 | 98.7297 | pan_paniscus | ENSPPAP00000018159 | 99.4354 | 98.7297 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 98.8003 | 97.6006 | pongo_abelii | ENSPPYP00000007694 | 98.8003 | 97.6006 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 99.1531 | 98.518 | pan_troglodytes | ENSPTRP00000012754 | 99.1531 | 98.518 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 99.3649 | 98.518 | gorilla_gorilla | ENSGGOP00000007548 | 99.5053 | 98.6572 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 71.4185 | 60.5505 | ornithorhynchus_anatinus | ENSOANP00000035626 | 72.2341 | 61.242 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 77.2759 | 65.7728 | sarcophilus_harrisii | ENSSHAP00000009227 | 75.7785 | 64.4983 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 72.1948 | 61.5385 | notamacropus_eugenii | ENSMEUP00000006633 | 74.5084 | 63.5106 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 74.1002 | 68.1722 | choloepus_hoffmanni | ENSCHOP00000005548 | 75.8671 | 69.7977 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 87.3677 | 81.6514 | dasypus_novemcinctus | ENSDNOP00000007879 | 87.3677 | 81.6514 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 80.7339 | 73.2533 | erinaceus_europaeus | ENSEEUP00000001297 | 82.7187 | 75.0542 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.5385 | 55.3282 | echinops_telfairi | ENSETEP00000012170 | 61.279 | 55.0949 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 95.9068 | 92.9428 | callithrix_jacchus | ENSCJAP00000046369 | 96.0424 | 93.0742 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 97.4594 | 95.9774 | cercocebus_atys | ENSCATP00000035511 | 97.5283 | 96.0452 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 97.6711 | 95.9774 | macaca_fascicularis | ENSMFAP00000022593 | 97.7401 | 96.0452 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 97.7417 | 96.1186 | macaca_mulatta | ENSMMUP00000025651 | 97.8107 | 96.1864 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 92.8017 | 90.5434 | macaca_nemestrina | ENSMNEP00000014157 | 95.6364 | 93.3091 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 97.6711 | 96.1186 | papio_anubis | ENSPANP00000014473 | 97.7401 | 96.1864 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 95.1306 | 93.5074 | mandrillus_leucophaeus | ENSMLEP00000017525 | 96.8391 | 95.1868 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 89.9788 | 84.3331 | tursiops_truncatus | ENSTTRP00000015530 | 90.4897 | 84.8119 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 87.8617 | 82.0748 | vulpes_vulpes | ENSVVUP00000008524 | 87.6761 | 81.9014 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 85.4622 | 76.6408 | mus_spretus | MGP_SPRETEiJ_P0083743 | 85.5226 | 76.6949 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 89.626 | 84.0508 | equus_asinus | ENSEASP00005009632 | 87.5258 | 82.0813 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 88.4263 | 81.7925 | capra_hircus | ENSCHIP00000006552 | 87.6837 | 81.1057 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 87.7911 | 81.1574 | panthera_leo | ENSPLOP00000025339 | 77.5078 | 71.6511 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 88.4263 | 82.0042 | ailuropoda_melanoleuca | ENSAMEP00000014621 | 88.0534 | 81.6585 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 89.132 | 82.6394 | bos_indicus_hybrid | ENSBIXP00005014678 | 88.3217 | 81.8881 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 84.7565 | 75.4411 | oryctolagus_cuniculus | ENSOCUP00000012010 | 84.9363 | 75.6011 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 90.4728 | 85.18 | equus_caballus | ENSECAP00000068873 | 88.292 | 83.1267 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 88.0734 | 82.2865 | canis_lupus_familiaris | ENSCAFP00845003429 | 87.8873 | 82.1127 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 88.0028 | 81.2279 | panthera_pardus | ENSPPRP00000007220 | 88.1272 | 81.3428 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 89.4848 | 82.7805 | marmota_marmota_marmota | ENSMMMP00000021885 | 90.0568 | 83.3097 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 90.8257 | 85.5328 | balaenoptera_musculus | ENSBMSP00010003126 | 90.8257 | 85.5328 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 86.4502 | 78.1228 | cavia_porcellus | ENSCPOP00000001592 | 87.1886 | 78.79 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 88.144 | 81.2279 | felis_catus | ENSFCAP00000005659 | 87.465 | 80.6022 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 77.4876 | 66.7608 | monodelphis_domestica | ENSMODP00000024262 | 75.672 | 65.1964 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 89.3437 | 82.9922 | bison_bison_bison | ENSBBBP00000015864 | 89.3437 | 82.9922 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 90.8963 | 85.3917 | monodon_monoceros | ENSMMNP00015009778 | 89.6936 | 84.2618 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 90.1905 | 83.5568 | sus_scrofa | ENSSSCP00000001986 | 89.2458 | 82.6816 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 91.7431 | 86.3091 | microcebus_murinus | ENSMICP00000040552 | 91.6784 | 86.2482 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 64.0085 | 57.9393 | octodon_degus | ENSODEP00000020850 | 89.891 | 81.3677 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.6796 | 57.8687 | jaculus_jaculus | ENSJJAP00000013641 | 88.4615 | 82.9959 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 88.144 | 81.7219 | ovis_aries_rambouillet | ENSOARP00020022294 | 87.4038 | 81.0357 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 88.2851 | 82.0748 | ursus_maritimus | ENSUMAP00000012377 | 87.6664 | 81.4996 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 74.3825 | 65.561 | ochotona_princeps | ENSOPRP00000010666 | 75.7728 | 66.7865 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 90.8257 | 85.1094 | delphinapterus_leucas | ENSDLEP00000010586 | 90.063 | 84.3947 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 90.7551 | 85.3917 | physeter_catodon | ENSPCTP00005031450 | 89.8672 | 84.5563 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 84.686 | 75.7939 | mus_caroli | MGP_CAROLIEiJ_P0080394 | 84.9858 | 76.0623 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 85.18 | 76.5702 | mus_musculus | ENSMUSP00000127995 | 85.0599 | 76.4623 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 64.2202 | 59.4213 | dipodomys_ordii | ENSDORP00000010369 | 88.0077 | 81.4313 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 73.8179 | 69.1602 | mustela_putorius_furo | ENSMPUP00000015547 | 85.6675 | 80.2621 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 96.048 | 93.1545 | aotus_nancymaae | ENSANAP00000042251 | 96.1837 | 93.2862 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 86.662 | 83.3451 | saimiri_boliviensis_boliviensis | ENSSBOP00000029255 | 95.6386 | 91.9782 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 87.0148 | 80.8045 | vicugna_pacos | ENSVPAP00000000078 | 87.1994 | 80.976 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 97.3888 | 95.6951 | chlorocebus_sabaeus | ENSCSAP00000013082 | 96.9782 | 95.2916 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 65.067 | 59.9153 | tupaia_belangeri | ENSTBEP00000005799 | 66.8116 | 61.5217 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 85.9562 | 78.5462 | mesocricetus_auratus | ENSMAUP00000007271 | 86.0777 | 78.6572 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 90.614 | 84.8977 | phocoena_sinus | ENSPSNP00000001522 | 89.7902 | 84.1259 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 90.2611 | 84.0508 | camelus_dromedarius | ENSCDRP00005017754 | 89.5032 | 83.345 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 92.3077 | 87.0148 | carlito_syrichta | ENSTSYP00000011350 | 92.569 | 87.2611 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 87.7205 | 81.0162 | panthera_tigris_altaica | ENSPTIP00000017901 | 87.7205 | 81.0162 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 84.2625 | 75.1588 | rattus_norvegicus | ENSRNOP00000015065 | 85.1641 | 75.9629 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 92.1665 | 86.7325 | prolemur_simus | ENSPSMP00000017103 | 92.2316 | 86.7938 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 85.0388 | 76.9231 | microtus_ochrogaster | ENSMOCP00000017935 | 86.1329 | 77.9128 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 73.1122 | 66.2668 | sorex_araneus | ENSSARP00000005834 | 78.3661 | 71.0287 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 86.0974 | 78.2639 | peromyscus_maniculatus_bairdii | ENSPEMP00000018930 | 86.3411 | 78.4855 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 89.3437 | 82.9217 | bos_mutus | ENSBMUP00000009536 | 88.5315 | 82.1678 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 84.9682 | 76.2174 | mus_spicilegus | ENSMSIP00000003472 | 85.0883 | 76.3251 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 86.0268 | 78.8285 | cricetulus_griseus_chok1gshd | ENSCGRP00001028072 | 86.4539 | 79.2199 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 73.4651 | 67.3959 | pteropus_vampyrus | ENSPVAP00000010975 | 73.673 | 67.5867 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 87.4382 | 80.6634 | myotis_lucifugus | ENSMLUP00000013381 | 87.8723 | 81.0638 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 77.4171 | 66.4079 | vombatus_ursinus | ENSVURP00010008414 | 75.7074 | 64.9413 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 87.156 | 79.5342 | rhinolophus_ferrumequinum | ENSRFEP00010031107 | 87.156 | 79.5342 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 87.3677 | 81.7219 | neovison_vison | ENSNVIP00000021144 | 87.4912 | 81.8375 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 81.4397 | 75.3705 | bos_taurus | ENSBTAP00000027057 | 87.6234 | 81.0934 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 63.3733 | 56.7396 | heterocephalus_glaber_female | ENSHGLP00000018471 | 85.5238 | 76.5714 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 94.8483 | 92.8017 | rhinopithecus_bieti | ENSRBIP00000011579 | 96.2751 | 94.1977 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 88.0734 | 82.4982 | canis_lupus_dingo | ENSCAFP00020020369 | 87.8873 | 82.3239 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 91.108 | 84.6154 | sciurus_vulgaris | ENSSVLP00005009481 | 90.7876 | 84.3179 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 77.3465 | 65.9139 | phascolarctos_cinereus | ENSPCIP00000007918 | 75.5862 | 64.4138 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 66.1962 | 61.115 | procavia_capensis | ENSPCAP00000008420 | 66.4777 | 61.3749 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 88.144 | 81.0868 | nannospalax_galili | ENSNGAP00000001749 | 88.5188 | 81.4316 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 89.3437 | 82.9217 | bos_grunniens | ENSBGRP00000014900 | 88.5315 | 82.1678 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 87.2265 | 79.8165 | chinchilla_lanigera | ENSCLAP00000017662 | 88.0342 | 80.5556 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 89.2025 | 82.0748 | cervus_hanglu_yarkandensis | ENSCHYP00000028132 | 88.3916 | 81.3287 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 88.3557 | 81.3691 | catagonus_wagneri | ENSCWAP00000016122 | 88.2311 | 81.2544 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 89.9788 | 83.2039 | ictidomys_tridecemlineatus | ENSSTOP00000004496 | 90.1697 | 83.3805 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 86.5208 | 79.1814 | otolemur_garnettii | ENSOGAP00000005104 | 87.5089 | 80.0857 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 94.7071 | 91.3903 | cebus_imitator | ENSCCAP00000010639 | 95.7887 | 92.434 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 90.1905 | 83.4862 | urocitellus_parryii | ENSUPAP00010018961 | 90.2542 | 83.5452 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 89.0614 | 81.7925 | moschus_moschiferus | ENSMMSP00000031505 | 89.0614 | 81.7925 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 96.6831 | 94.9188 | rhinopithecus_roxellana | ENSRROP00000028177 | 96.7514 | 94.9859 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 85.18 | 76.9231 | mus_pahari | MGP_PahariEiJ_P0017243 | 85.7244 | 77.4148 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 88.0028 | 83.0628 | propithecus_coquereli | ENSPCOP00000000241 | 92.2337 | 87.0562 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 33.0981 | 21.3832 | caenorhabditis_elegans | T04A11.6.1 | 47.4696 | 30.668 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 43.5427 | 27.0289 | drosophila_melanogaster | FBpp0081910 | 41.4929 | 25.7566 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 21.5243 | 14.9612 | ciona_intestinalis | ENSCINP00000036140 | 55.5556 | 38.6157 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 39.4495 | 30.6281 | petromyzon_marinus | ENSPMAP00000000495 | 75.1344 | 58.3333 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 30.1341 | 23.0064 | eptatretus_burgeri | ENSEBUP00000025579 | 61.1748 | 46.7049 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 48.8356 | 39.8024 | callorhinchus_milii | ENSCMIP00000044113 | 71.1934 | 58.0247 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 64.9259 | 51.8701 | latimeria_chalumnae | ENSLACP00000007317 | 63.4483 | 50.6897 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.6796 | 46.6478 | lepisosteus_oculatus | ENSLOCP00000017609 | 59.0142 | 44.632 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 10.374 | 5.29287 | clupea_harengus | ENSCHAP00000014529 | 45.3704 | 23.1481 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.2562 | 45.8716 | danio_rerio | ENSDARP00000153051 | 60.3197 | 45.1703 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.1856 | 47.0007 | carassius_auratus | ENSCARP00000107602 | 61.0134 | 46.8684 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.3387 | 46.2244 | astyanax_mexicanus | ENSAMXP00000017467 | 60.2962 | 46.1918 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 53.4227 | 41.9195 | ictalurus_punctatus | ENSIPUP00000000729 | 66.8728 | 52.4735 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.6916 | 46.4361 | esox_lucius | ENSELUP00000019791 | 58.3843 | 44.6707 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 57.9393 | 46.0127 | electrophorus_electricus | ENSEEEP00000035037 | 64.0906 | 50.8977 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 62.3148 | 47.2124 | oncorhynchus_kisutch | ENSOKIP00005015773 | 60.068 | 45.5102 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.8913 | 46.6478 | oncorhynchus_mykiss | ENSOMYP00000067552 | 61.0299 | 45.9986 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 62.8088 | 46.9301 | salmo_salar | ENSSSAP00000110484 | 60.5031 | 45.2073 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 62.6676 | 46.789 | salmo_trutta | ENSSTUP00000086462 | 60.1219 | 44.8883 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 14.9612 | 11.0092 | gadus_morhua | ENSGMOP00000011518 | 63.8554 | 46.988 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 21.5243 | 16.302 | gadus_morhua | ENSGMOP00000011634 | 64.482 | 48.8372 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 59.2802 | 42.9781 | gadus_morhua | ENSGMOP00000017886 | 61.2691 | 44.4201 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.6916 | 45.5893 | fundulus_heteroclitus | ENSFHEP00000001801 | 61.21 | 45.9786 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.3267 | 45.4481 | poecilia_reticulata | ENSPREP00000011311 | 62.1158 | 46.0329 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.4093 | 45.1658 | xiphophorus_maculatus | ENSXMAP00000027372 | 61.2303 | 45.7797 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.8327 | 46.0127 | oryzias_latipes | ENSORLP00000027048 | 61.8808 | 46.8055 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 59.0685 | 44.6718 | cyclopterus_lumpus | ENSCLMP00005016371 | 61.7257 | 46.6814 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.4679 | 45.3776 | oreochromis_niloticus | ENSONIP00000019124 | 61.9048 | 45.7001 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.1856 | 45.6598 | haplochromis_burtoni | ENSHBUP00000014122 | 61.7082 | 46.0498 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.5505 | 45.0953 | astatotilapia_calliptera | ENSACLP00000030031 | 61.6379 | 45.9052 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 59.3507 | 46.1538 | sparus_aurata | ENSSAUP00010051335 | 58.6881 | 45.6385 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.9739 | 46.6478 | lates_calcarifer | ENSLCAP00010010683 | 61.1465 | 46.7799 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 62.8793 | 49.6119 | xenopus_tropicalis | ENSXETP00000017154 | 65.1316 | 51.3889 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.609 | 48.6239 | chrysemys_picta_bellii | ENSCPBP00000036113 | 68.7943 | 54.2947 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 56.7396 | 44.813 | sphenodon_punctatus | ENSSPUP00000000081 | 73.3577 | 57.938 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 67.2548 | 52.5759 | crocodylus_porosus | ENSCPRP00005002864 | 65.0068 | 50.8186 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 62.3148 | 47.6358 | laticauda_laticaudata | ENSLLTP00000022373 | 62.2269 | 47.5687 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 23.3592 | 19.9012 | notechis_scutatus | ENSNSUP00000028494 | 81.9307 | 69.802 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 45.0953 | 38.391 | pseudonaja_textilis | ENSPTXP00000022384 | 83.6387 | 71.2042 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.3973 | 48.765 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000006514 | 66.8203 | 53.0722 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 63.9379 | 51.0233 | gallus_gallus | ENSGALP00010022807 | 65.6522 | 52.3913 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 59.139 | 47.2124 | meleagris_gallopavo | ENSMGAP00000010882 | 66.7197 | 53.2643 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 56.1044 | 45.1658 | serinus_canaria | ENSSCAP00000017987 | 65.2174 | 52.5021 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 53.3522 | 43.1193 | parus_major | ENSPMJP00000015341 | 64.6154 | 52.2222 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 62.9499 | 49.753 | anolis_carolinensis | ENSACAP00000025027 | 63.1728 | 49.9292 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.9033 | 45.6598 | neolamprologus_brichardi | ENSNBRP00000025793 | 62.4909 | 46.8501 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 57.8687 | 44.6013 | naja_naja | ENSNNAP00000022094 | 64.7709 | 49.921 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 52.9287 | 43.5427 | podarcis_muralis | ENSPMRP00000033013 | 73.2422 | 60.2539 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 54.4107 | 44.5307 | geospiza_fortis | ENSGFOP00000019164 | 71.3228 | 58.3719 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 58.2216 | 47.283 | taeniopygia_guttata | ENSTGUP00000010286 | 63.4615 | 51.5385 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 62.8793 | 47.0713 | erpetoichthys_calabaricus | ENSECRP00000030972 | 60.6122 | 45.3741 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 59.4213 | 44.813 | kryptolebias_marmoratus | ENSKMAP00000022385 | 60.8382 | 45.8815 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.9619 | 47.3536 | salvator_merianae | ENSSMRP00000027994 | 63.6232 | 48.6232 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.9739 | 45.9421 | oryzias_javanicus | ENSOJAP00000031090 | 61.8468 | 46.5999 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.621 | 44.813 | amphilophus_citrinellus | ENSACIP00000030785 | 62.2915 | 46.0479 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.5505 | 45.307 | dicentrarchus_labrax | ENSDLAP00005029172 | 60.2105 | 45.0526 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.3387 | 45.2364 | amphiprion_percula | ENSAPEP00000025450 | 59.4576 | 44.5758 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.9033 | 46.0127 | oryzias_sinensis | ENSOSIP00000015758 | 61.9526 | 46.8055 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 35.7092 | 30.7692 | pelodiscus_sinensis | ENSPSIP00000010827 | 90.8438 | 78.2765 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 55.4693 | 44.8836 | gopherus_evgoodei | ENSGEVP00005015160 | 70.3671 | 56.9382 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 55.2576 | 44.813 | chelonoidis_abingdonii | ENSCABP00000010085 | 74.7851 | 60.6495 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.3387 | 45.5187 | seriola_dumerili | ENSSDUP00000014998 | 61.4224 | 46.3362 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 58.2922 | 43.5427 | cottoperca_gobio | ENSCGOP00000032574 | 59.639 | 44.5487 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 62.7382 | 49.259 | struthio_camelus_australis | ENSSCUP00000017735 | 69.1291 | 54.2768 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 65.9139 | 52.1524 | anser_brachyrhynchus | ENSABRP00000007605 | 65.0871 | 51.4983 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 59.0685 | 44.319 | gasterosteus_aculeatus | ENSGACP00000014383 | 61.5441 | 46.1765 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 56.2456 | 43.1193 | cynoglossus_semilaevis | ENSCSEP00000021128 | 61.3077 | 47 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.7622 | 45.4481 | poecilia_formosa | ENSPFOP00000005199 | 63.0307 | 47.1449 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.4093 | 45.7304 | myripristis_murdjan | ENSMMDP00005001078 | 60.5803 | 45.8599 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 59.4213 | 44.6718 | sander_lucioperca | ENSSLUP00000020090 | 61.3256 | 46.1034 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 54.4107 | 44.813 | strigops_habroptila | ENSSHBP00005009212 | 72.5306 | 59.7366 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.2682 | 45.1658 | amphiprion_ocellaris | ENSAOCP00000019728 | 62.064 | 46.5116 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 58.2216 | 45.5893 | terrapene_carolina_triunguis | ENSTMTP00000014367 | 68.3513 | 53.5211 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.9619 | 46.8596 | hucho_hucho | ENSHHUP00000046780 | 60.2608 | 45.5731 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.8327 | 45.8716 | betta_splendens | ENSBSLP00000049183 | 61.3523 | 46.2633 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 59.9859 | 46.8596 | pygocentrus_nattereri | ENSPNAP00000005509 | 60.5413 | 47.2934 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.4799 | 45.4481 | anabas_testudineus | ENSATEP00000009122 | 62.282 | 46.8023 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 23.1475 | 15.5963 | ciona_savignyi | ENSCSAVP00000012803 | 55.8773 | 37.6491 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 39.6613 | 22.3712 | saccharomyces_cerevisiae | YMR190C | 38.839 | 21.9074 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.4799 | 45.7304 | seriola_lalandi_dorsalis | ENSSLDP00000000679 | 61.6104 | 46.5852 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 45.0953 | 36.1327 | labrus_bergylta | ENSLBEP00000024461 | 75.5319 | 60.5201 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 55.5399 | 39.379 | pundamilia_nyererei | ENSPNYP00000014484 | 58.7752 | 41.6729 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.6916 | 46.5773 | mastacembelus_armatus | ENSMAMP00000012848 | 61.6046 | 47.2779 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.6916 | 45.8716 | stegastes_partitus | ENSSPAP00000018586 | 61.21 | 46.2633 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 62.8088 | 47.2124 | oncorhynchus_tshawytscha | ENSOTSP00005047792 | 60.462 | 45.4484 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 59.7036 | 44.813 | acanthochromis_polyacanthus | ENSAPOP00000015041 | 60.1707 | 45.1636 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 63.0205 | 50.1764 | aquila_chrysaetos_chrysaetos | ENSACCP00020003599 | 66.6916 | 53.0993 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 57.7276 | 42.7664 | nothobranchius_furzeri | ENSNFUP00015015696 | 50.0306 | 37.0642 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 42.0607 | 29.0755 | cyprinodon_variegatus | ENSCVAP00000014694 | 54.6288 | 37.7635 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 59.9859 | 46.0833 | denticeps_clupeoides | ENSDCDP00000015862 | 59.775 | 45.9212 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 59.9153 | 45.1658 | larimichthys_crocea | ENSLCRP00005055960 | 60.5132 | 45.6165 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 57.657 | 43.4721 | takifugu_rubripes | ENSTRUP00000078695 | 58.6083 | 44.1894 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 54.1284 | 44.319 | ficedula_albicollis | ENSFALP00000011477 | 70.0457 | 57.3516 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 49.753 | 39.8024 | hippocampus_comes | ENSHCOP00000023146 | 72.9814 | 58.3851 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.1856 | 46.3656 | cyprinus_carpio_carpio | ENSCCRP00000117094 | 60.2502 | 45.6567 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 59.7036 | 45.307 | cyprinus_carpio_carpio | ENSCCRP00000154670 | 58.5062 | 44.3983 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 62.597 | 49.8236 | coturnix_japonica | ENSCJPP00005027231 | 64.6031 | 51.4202 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.7622 | 45.4481 | poecilia_latipinna | ENSPLAP00000028184 | 63.0307 | 47.1449 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 27.9464 | 21.5243 | sinocyclocheilus_grahami | ENSSGRP00000064987 | 66.3317 | 51.0888 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 56.175 | 43.0487 | sinocyclocheilus_grahami | ENSSGRP00000089578 | 61.1367 | 46.851 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.621 | 45.307 | maylandia_zebra | ENSMZEP00005029946 | 62.2013 | 46.4881 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 58.2216 | 43.825 | tetraodon_nigroviridis | ENSTNIP00000022447 | 61.6592 | 46.4126 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.5505 | 46.8596 | paramormyrops_kingsleyae | ENSPKIP00000026501 | 59.5007 | 46.0472 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 61.8913 | 48.4121 | leptobrachium_leishanense | ENSLLEP00000024839 | 60.7762 | 47.5398 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.9739 | 45.801 | scophthalmus_maximus | ENSSMAP00000064008 | 61.9355 | 46.5233 |
| ENSG00000197299 | homo_sapiens | ENSP00000347232 | 60.4799 | 45.1658 | oryzias_melastigma | ENSOMEP00000036003 | 62.6462 | 46.7836 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000079 | involved_in regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0000228 | located_in nuclear chromosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0000400 | enables four-way junction DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0000403 | enables Y-form DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0000405 | enables bubble DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0000723 | involved_in telomere maintenance | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0000724 | involved_in double-strand break repair via homologous recombination | 3 | IBA,IDA,NAS | Homo_sapiens(9606) | Process |
| GO:0000729 | involved_in DNA double-strand break processing | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0000781 | colocalizes_with chromosome, telomeric region | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0000800 | located_in lateral element | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0002039 | enables p53 binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0003677 | enables DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003678 | enables DNA helicase activity | 2 | IDA,IMP | Homo_sapiens(9606) | Function |
| GO:0003697 | enables single-stranded DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0004386 | enables helicase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005524 | enables ATP binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 3 | IBA,IDA,NAS | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005657 | located_in replication fork | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0005694 | is_active_in chromosome | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0005730 | located_in nucleolus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0006260 | involved_in DNA replication | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0006268 | involved_in DNA unwinding involved in DNA replication | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0006281 | involved_in DNA repair | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0006310 | involved_in DNA recombination | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0006974 | involved_in DNA damage response | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0007095 | involved_in mitotic G2 DNA damage checkpoint signaling | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0008094 | enables ATP-dependent activity, acting on DNA | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0008270 | enables zinc ion binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0009378 | enables four-way junction helicase activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0010165 | involved_in response to X-ray | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0016363 | located_in nuclear matrix | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0016605 | colocalizes_with PML body | 2 | IDA | Homo_sapiens(9606) | Component |
| GO:0016887 | enables ATP hydrolysis activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0031297 | involved_in replication fork processing | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0031422 | part_of RecQ family helicase-topoisomerase III complex | 1 | IPI | Homo_sapiens(9606) | Component |
| GO:0032201 | involved_in telomere maintenance via semi-conservative replication | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0032508 | involved_in DNA duplex unwinding | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032991 | part_of protein-containing complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0042802 | enables identical protein binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0042803 | enables protein homodimerization activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0043138 | enables 3-5 DNA helicase activity | 2 | IBA,TAS | Homo_sapiens(9606) | Function |
| GO:0044806 | involved_in G-quadruplex DNA unwinding | 3 | IBA,IDA,ISS | Homo_sapiens(9606) | Process |
| GO:0045893 | involved_in positive regulation of DNA-templated transcription | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045910 | involved_in negative regulation of DNA recombination | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0051259 | involved_in protein complex oligomerization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051260 | involved_in protein homooligomerization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051782 | involved_in negative regulation of cell division | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0051880 | enables G-quadruplex DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0061749 | enables forked DNA-dependent helicase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0061820 | involved_in telomeric D-loop disassembly | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0061821 | enables telomeric D-loop binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0061849 | enables telomeric G-quadruplex DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0071139 | involved_in resolution of recombination intermediates | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071479 | involved_in cellular response to ionizing radiation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0072711 | involved_in cellular response to hydroxyurea | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0072757 | involved_in cellular response to camptothecin | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0090329 | involved_in regulation of DNA-templated DNA replication | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0090656 | involved_in t-circle formation | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0140677 | enables molecular function activator activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:1905773 | enables 8-hydroxy-2-deoxyguanosine DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:1990814 | enables DNA/DNA annealing activity | 1 | IDA | Homo_sapiens(9606) | Function |