Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | CancerSARS-COV-OC43FlavivirusesSARS-COV-2DengueZika |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | N.A. |
| Localizations | Stress granucle |
| BulkPerturb-seq | N.A. |
Description
| Ensembl ID | ENSG00000183684 | Gene ID | 10189 | Accession | 19071 |
| Symbol | ALYREF | Alias | ALY;BEF;REF;THOC4;ALY/REF | Full Name | Aly/REF export factor |
| Status | Confidence | Length | 3752 bases | Strand | Minus strand |
| Position | 17 : 81887835 - 81891586 | RNA binding domain | RRM_1 | ||
| Summary | The protein encoded by this gene is a heat stable, nuclear protein and functions as a molecular chaperone. It is thought to regulate dimerization, DNA binding, and transcriptional activity of basic region-leucine zipper (bZIP) proteins. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000421592 | RRM_1 | PF00076 | 9.9e-13 | 1 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 35886072 | The Epitranscriptome in miRNAs: Crosstalk, Detection, and Function in Cancer. | Daniel D Del | 2022-07-21 | Genes |
| 28927264 | Comparative Interactomes of VRK1 and VRK3 with Their Distinct Roles in the Cell Cycle of Liver Cancer. | Namgyu Lee | 2017-09-30 | Molecules and cells |
| 30858280 | ALYREF links 3'-end processing to nuclear export of non-polyadenylated mRNAs. | Jing Fan | 2019-05-02 | The EMBO journal |
| 35669236 | Systematic evaluation of parameters in RNA bisulfite sequencing data generation and analysis. | Zachary Johnson | 2022-06-01 | NAR genomics and bioinformatics |
| 36934090 | ALYREF mediates RNA m5C modification to promote hepatocellular carcinoma progression. | Chen Xue | 2023-03-18 | Signal transduction and targeted therapy |
| 37393317 | NSUN2 stimulates tumor progression via enhancing TIAM2 mRNA stability in pancreatic cancer. | Guizhen Zhang | 2023-07-01 | Cell death discovery |
| 28934468 | ALYREF mainly binds to the 5' and the 3' regions of the mRNA in vivo. | Min Shi | 2017-09-19 | Nucleic acids research |
| 35776213 | ALYREF, a novel factor involved in breast carcinogenesis, acts through transcriptional and post-transcriptional mechanisms selectively regulating the short NEAT1 isoform. | Christiane Klec | 2022-07-01 | Cellular and molecular life sciences : CMLS |
| 32440736 | Mapping the epigenetic modifications of DNA and RNA. | Lin-Yong Zhao | 2020-11-01 | Protein & cell |
| 24550725 | Competitive and cooperative interactions mediate RNA transfer from herpesvirus saimiri ORF57 to the mammalian export adaptor ALYREF. | B Richard Tunnicliffe | 2014-02-01 | PLoS pathogens |
| 26773052 | A short conserved motif in ALYREF directs cap- and EJC-dependent assembly of export complexes on spliced mRNAs. | M Agnieszka Gromadzka | 2016-03-18 | Nucleic acids research |
| 36389669 | RNA m5C regulator-mediated modification patterns and the cross-talk between tumor microenvironment infiltration in gastric cancer. | Qiang Zhang | 2022-01-01 | Frontiers in immunology |
| 36690199 | Quadruplexes and aging: G4-binding proteins regulate the presence of miRNA in small extracellular vesicles (sEVs). | Václav Brázda | 2023-01-20 | Biochimie |
| 35210361 | 5-methylcytosine modification by Plasmodium NSUN2 stabilizes mRNA and mediates the development of gametocytes. | Meng Liu | 2022-03-01 | Proceedings of the National Academy of Sciences of the United States of America |
| 34530450 | ZFC3H1 prevents RNA trafficking into nuclear speckles through condensation. | Yimin Wang | 2021-10-11 | Nucleic acids research |
| 27976729 | Interactomic analysis of REST/NRSF and implications of its functional links with the transcription suppressor TRIM28 during neuronal differentiation. | Namgyu Lee | 2016-12-15 | Scientific reports |
| 36240321 | 5-methylcytosine (m5C) RNA modification controls the innate immune response to virus infection by regulating type I interferons. | Yuexiu Zhang | 2022-10-18 | Proceedings of the National Academy of Sciences of the United States of America |
| 28585384 | Modeling the C9ORF72 repeat expansion mutation using human induced pluripotent stem cells. | T Bhuvaneish Selvaraj | 2017-07-01 | Brain pathology (Zurich, Switzerland) |
| 32748033 | Epigenetic Regulation of Endothelial Cell Function by Nucleic Acid Methylation in Cardiac Homeostasis and Disease. | Adam Russell-Hallinan | 2021-10-01 | Cardiovascular drugs and therapy |
| 31641093 | Molecular Mechanism of SR Protein Kinase 1 Inhibition by the Herpes Virus Protein ICP27. | B Richard Tunnicliffe | 2019-10-22 | mBio |
| 36899389 | Exosomal cargos-mediated metabolic reprogramming in tumor microenvironment. | Shiming Tan | 2023-03-10 | Journal of experimental & clinical cancer research : CR |
| 35688334 | Extracellular vesicles in cancer therapy. | Emily Shizhen Wang | 2022-11-01 | Seminars in cancer biology |
| 36437944 | The role of RNA modification in the generation of acquired drug resistance in glioma. | Yu Yan | 2022-01-01 | Frontiers in genetics |
| 33991457 | The role of the HIF-1α/ALYREF/PKM2 axis in glycolysis and tumorigenesis of bladder cancer. | Jing-Zi Wang | 2021-07-01 | Cancer communications (London, England) |
| 30218090 | The m6A-methylase complex recruits TREX and regulates mRNA export. | Simon Lesbirel | 2018-09-14 | Scientific reports |
| 34630524 | Development and Validation of a Novel Gene Signature for Predicting the Prognosis by Identifying m5C Modification Subtypes of Cervical Cancer. | Jing Yu | 2021-01-01 | Frontiers in genetics |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| ALYREF-202 | ENST00000505490 | 1097 | 264aa | ENSP00000421592 |
| ALYREF-201 | ENST00000504015 | 254 | No protein | - |
| ALYREF-204 | ENST00000512673 | 441 | No protein | - |
| ALYREF-203 | ENST00000511412 | 533 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
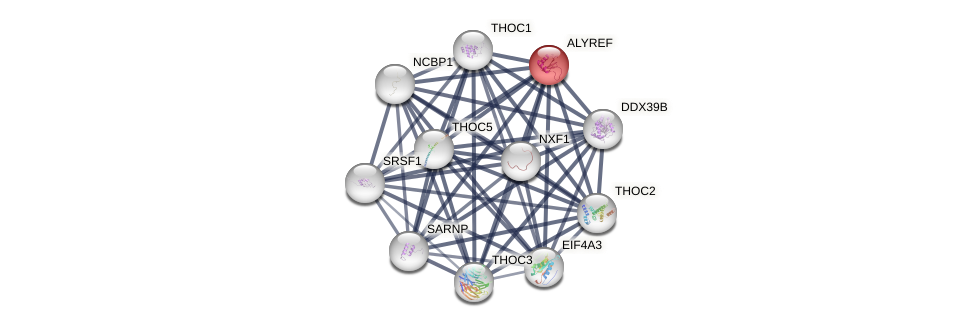
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000183684 | homo_sapiens | ENSP00000357683 | 36.2903 | 25.8065 | homo_sapiens | ENSP00000421592 | 34.0909 | 24.2424 |
| ENSG00000183684 | homo_sapiens | ENSP00000252115 | 27.0784 | 17.1021 | homo_sapiens | ENSP00000421592 | 43.1818 | 27.2727 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.4848 | 98.1061 | nomascus_leucogenys | ENSNLEP00000041346 | 91.2281 | 90.8772 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 99.2424 | 99.2424 | pongo_abelii | ENSPPYP00000009820 | 99.2424 | 99.2424 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.4848 | 98.1061 | pan_troglodytes | ENSPTRP00000089578 | 91.2281 | 90.8772 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.8636 | 98.8636 | gorilla_gorilla | ENSGGOP00000013592 | 95.2555 | 95.2555 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 87.5 | 85.2273 | ornithorhynchus_anatinus | ENSOANP00000046142 | 75.9868 | 74.0132 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 95.4545 | 93.9394 | sarcophilus_harrisii | ENSSHAP00000030588 | 97.6744 | 96.124 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 61.3636 | 59.8485 | notamacropus_eugenii | ENSMEUP00000002015 | 70.7424 | 68.9956 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 71.5909 | 68.5606 | dasypus_novemcinctus | ENSDNOP00000001990 | 79.7468 | 76.3713 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 63.6364 | 63.6364 | erinaceus_europaeus | ENSEEUP00000009246 | 98.2456 | 98.2456 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 63.6364 | 61.7424 | echinops_telfairi | ENSETEP00000013465 | 95.4545 | 92.6136 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.4848 | 98.4848 | callithrix_jacchus | ENSCJAP00000064668 | 98.4848 | 98.4848 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 99.6212 | 98.1061 | cercocebus_atys | ENSCATP00000018739 | 90.6897 | 89.3103 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 87.5 | 86.7424 | macaca_fascicularis | ENSMFAP00000008167 | 87.8327 | 87.0722 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 99.6212 | 98.1061 | macaca_mulatta | ENSMMUP00000042995 | 90.6897 | 89.3103 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 99.6212 | 98.1061 | macaca_nemestrina | ENSMNEP00000041705 | 90.6897 | 89.3103 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 99.6212 | 99.2424 | papio_anubis | ENSPANP00000016367 | 99.6212 | 99.2424 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 61.7424 | 60.6061 | mandrillus_leucophaeus | ENSMLEP00000038608 | 78.744 | 77.2947 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 95.0758 | 93.5606 | tursiops_truncatus | ENSTTRP00000011643 | 98.4314 | 96.8627 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 94.697 | 93.1818 | mus_spretus | MGP_SPRETEiJ_P0033025 | 98.0392 | 96.4706 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 76.8939 | 74.2424 | mus_spretus | MGP_SPRETEiJ_P0022297 | 93.1193 | 89.9083 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 96.5909 | 95.4545 | capra_hircus | ENSCHIP00000017673 | 95.5056 | 94.382 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 68.9394 | 67.4242 | loxodonta_africana | ENSLAFP00000002571 | 97.3262 | 95.1872 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.1061 | 96.9697 | panthera_leo | ENSPLOP00000020317 | 98.4791 | 97.3384 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 71.2121 | 67.0455 | ailuropoda_melanoleuca | ENSAMEP00000018449 | 57.8462 | 54.4615 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.4848 | 97.3485 | bos_indicus_hybrid | ENSBIXP00005010529 | 98.8593 | 97.7186 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.4848 | 97.3485 | equus_caballus | ENSECAP00000025726 | 98.8593 | 97.7186 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.4848 | 96.5909 | canis_lupus_familiaris | ENSCAFP00845012529 | 98.8593 | 96.9582 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.1061 | 96.9697 | panthera_pardus | ENSPPRP00000019478 | 98.4791 | 97.3384 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.4848 | 97.3485 | balaenoptera_musculus | ENSBMSP00010023735 | 98.8593 | 97.7186 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 95.8333 | 94.3182 | cavia_porcellus | ENSCPOP00000026783 | 98.8281 | 97.2656 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.1061 | 96.9697 | felis_catus | ENSFCAP00000029154 | 98.4791 | 97.3384 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 62.8788 | 57.9545 | ursus_americanus | ENSUAMP00000021563 | 65.6126 | 60.4743 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.1061 | 96.2121 | monodelphis_domestica | ENSMODP00000003678 | 97.7358 | 95.8491 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 70.4545 | 68.1818 | bison_bison_bison | ENSBBBP00000007650 | 78.1513 | 75.6302 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 96.5909 | 95.0758 | monodon_monoceros | ENSMMNP00015026356 | 99.2218 | 97.6654 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 77.6515 | 72.3485 | sus_scrofa | ENSSSCP00000071154 | 58.908 | 54.8851 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 97.7273 | 96.5909 | microcebus_murinus | ENSMICP00000015851 | 94.1606 | 93.0657 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 96.5909 | 94.697 | octodon_degus | ENSODEP00000011236 | 98.0769 | 96.1538 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 93.5606 | 92.0455 | jaculus_jaculus | ENSJJAP00000002422 | 96.8627 | 95.2941 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.4848 | 97.3485 | ovis_aries_rambouillet | ENSOARP00020032834 | 98.8593 | 97.7186 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 60.9848 | 57.5758 | ursus_maritimus | ENSUMAP00000022412 | 64.6586 | 61.0442 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 53.4091 | 51.8939 | ochotona_princeps | ENSOPRP00000005085 | 55.0781 | 53.5156 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 95.4545 | 93.5606 | delphinapterus_leucas | ENSDLEP00000004273 | 91.9708 | 90.146 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.4848 | 97.3485 | physeter_catodon | ENSPCTP00005030518 | 84.4156 | 83.4416 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 94.697 | 93.1818 | mus_caroli | MGP_CAROLIEiJ_P0031574 | 98.0392 | 96.4706 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 75.7576 | 73.4848 | mus_caroli | MGP_CAROLIEiJ_P0021222 | 89.6861 | 86.9955 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 77.2727 | 74.6212 | mus_musculus | ENSMUSP00000080242 | 93.578 | 90.367 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 94.697 | 93.1818 | mus_musculus | ENSMUSP00000026125 | 98.0392 | 96.4706 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 71.5909 | 68.9394 | dipodomys_ordii | ENSDORP00000019908 | 77.1429 | 74.2857 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 87.1212 | 86.3636 | aotus_nancymaae | ENSANAP00000020895 | 77.7027 | 77.027 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 99.6212 | 99.2424 | chlorocebus_sabaeus | ENSCSAP00000015818 | 99.6212 | 99.2424 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 64.3939 | 62.8788 | tupaia_belangeri | ENSTBEP00000005011 | 95.5056 | 93.2584 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 79.5455 | 76.5152 | mesocricetus_auratus | ENSMAUP00000023396 | 86.7769 | 83.4711 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.4848 | 96.9697 | phocoena_sinus | ENSPSNP00000018916 | 98.8593 | 97.3384 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 84.8485 | 83.7121 | camelus_dromedarius | ENSCDRP00005013094 | 92.9461 | 91.7012 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 68.1818 | 67.0455 | carlito_syrichta | ENSTSYP00000015532 | 69.2308 | 68.0769 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 63.2576 | 58.3333 | panthera_tigris_altaica | ENSPTIP00000003766 | 58.1882 | 53.6585 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 95.0758 | 93.5606 | rattus_norvegicus | ENSRNOP00000066841 | 98.0469 | 96.4844 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 97.7273 | 95.8333 | prolemur_simus | ENSPSMP00000018698 | 94.1606 | 92.3358 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 83.3333 | 81.4394 | microtus_ochrogaster | ENSMOCP00000018538 | 85.6031 | 83.6576 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 94.3182 | 92.803 | mus_spicilegus | ENSMSIP00000000254 | 97.6471 | 96.0784 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 76.5152 | 74.2424 | mus_spicilegus | ENSMSIP00000028563 | 92.6606 | 89.9083 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 93.9394 | 91.6667 | cricetulus_griseus_chok1gshd | ENSCGRP00001018443 | 97.2549 | 94.902 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 77.2727 | 75.3788 | pteropus_vampyrus | ENSPVAP00000016855 | 80.315 | 78.3465 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 70.0758 | 60.2273 | myotis_lucifugus | ENSMLUP00000020153 | 64.4599 | 55.4007 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 67.4242 | 57.5758 | myotis_lucifugus | ENSMLUP00000019626 | 60.7509 | 51.8771 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 64.7727 | 53.7879 | myotis_lucifugus | ENSMLUP00000013699 | 72.1519 | 59.9156 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 66.6667 | 55.6818 | myotis_lucifugus | ENSMLUP00000009221 | 61.9718 | 51.7606 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 53.7879 | 45.4545 | myotis_lucifugus | ENSMLUP00000021742 | 67.619 | 57.1429 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 68.9394 | 60.2273 | myotis_lucifugus | ENSMLUP00000021978 | 60.4651 | 52.8239 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 69.697 | 59.8485 | myotis_lucifugus | ENSMLUP00000022768 | 64.1115 | 55.0523 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 68.1818 | 59.0909 | myotis_lucifugus | ENSMLUP00000016883 | 59.8007 | 51.8272 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 65.5303 | 55.303 | myotis_lucifugus | ENSMLUP00000021472 | 60.2787 | 50.8711 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 73.1061 | 65.1515 | myotis_lucifugus | ENSMLUP00000021851 | 66.782 | 59.5156 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 58.3333 | 49.2424 | myotis_lucifugus | ENSMLUP00000006948 | 59.0038 | 49.8084 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 66.2879 | 58.3333 | myotis_lucifugus | ENSMLUP00000008613 | 59.5238 | 52.381 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 72.3485 | 63.6364 | myotis_lucifugus | ENSMLUP00000002383 | 74.6094 | 65.625 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 97.3485 | 94.697 | vombatus_ursinus | ENSVURP00010011246 | 87.415 | 85.034 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 96.9697 | 94.697 | rhinolophus_ferrumequinum | ENSRFEP00010008103 | 64.4836 | 62.9723 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 56.8182 | 51.8939 | neovison_vison | ENSNVIP00000020991 | 66.3717 | 60.6195 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 96.5909 | 95.4545 | bos_taurus | ENSBTAP00000011202 | 99.2218 | 98.0545 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 94.697 | 92.0455 | heterocephalus_glaber_female | ENSHGLP00000011561 | 97.6562 | 94.9219 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 70.0758 | 67.803 | rhinopithecus_bieti | ENSRBIP00000031666 | 75.2033 | 72.7642 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 96.5909 | 94.697 | canis_lupus_dingo | ENSCAFP00020012248 | 99.2218 | 97.2763 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 97.7273 | 96.2121 | sciurus_vulgaris | ENSSVLP00005022991 | 98.0989 | 96.5779 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 86.7424 | 83.7121 | phascolarctos_cinereus | ENSPCIP00000042019 | 44.8141 | 43.2485 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 80.303 | 78.4091 | procavia_capensis | ENSPCAP00000012288 | 83.7945 | 81.8182 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.4848 | 97.3485 | bos_grunniens | ENSBGRP00000006363 | 98.8593 | 97.7186 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 84.8485 | 82.5758 | chinchilla_lanigera | ENSCLAP00000021928 | 94.5148 | 91.9831 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 92.4242 | 90.9091 | cervus_hanglu_yarkandensis | ENSCHYP00000010370 | 93.8462 | 92.3077 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 95.4545 | 94.3182 | catagonus_wagneri | ENSCWAP00000027083 | 97.6744 | 96.5116 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 71.2121 | 69.697 | otolemur_garnettii | ENSOGAP00000011107 | 80.6867 | 78.97 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.8636 | 98.8636 | cebus_imitator | ENSCCAP00000005682 | 98.8636 | 98.8636 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 75.7576 | 73.8636 | cebus_imitator | ENSCCAP00000024898 | 92.5926 | 90.2778 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 95.4545 | 93.9394 | urocitellus_parryii | ENSUPAP00010000440 | 98.4375 | 96.875 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 98.4848 | 97.3485 | moschus_moschiferus | ENSMMSP00000031586 | 98.8593 | 97.7186 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 75.7576 | 72.7273 | rhinopithecus_roxellana | ENSRROP00000044878 | 71.6846 | 68.8172 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 94.697 | 93.1818 | mus_pahari | MGP_PahariEiJ_P0038287 | 98.0392 | 96.4706 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 97.7273 | 96.9697 | propithecus_coquereli | ENSPCOP00000018034 | 98.0989 | 97.3384 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 41.6667 | 26.8939 | caenorhabditis_elegans | F23B2.6.1 | 48.4581 | 31.2775 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 43.5606 | 28.0303 | caenorhabditis_elegans | M18.7a.1 | 47.9167 | 30.8333 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 82.197 | 78.0303 | latimeria_chalumnae | ENSLACP00000006417 | 84.1085 | 79.845 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 67.803 | 58.7121 | clupea_harengus | ENSCHAP00000057055 | 60.473 | 52.3649 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 80.303 | 74.6212 | electrophorus_electricus | ENSEEEP00000047183 | 77.9412 | 72.4265 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 79.1667 | 75.3788 | oryzias_latipes | ENSORLP00000015435 | 77.9851 | 74.2537 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 67.0455 | 58.7121 | cyclopterus_lumpus | ENSCLMP00005008746 | 68.6047 | 60.0775 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 78.7879 | 74.6212 | cyclopterus_lumpus | ENSCLMP00005010444 | 77.037 | 72.963 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 81.8182 | 77.6515 | sparus_aurata | ENSSAUP00010064697 | 79.4118 | 75.3676 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 81.4394 | 78.0303 | xenopus_tropicalis | ENSXETP00000060614 | 88.843 | 85.124 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 85.6061 | 82.5758 | sphenodon_punctatus | ENSSPUP00000005952 | 87.5969 | 84.4961 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 82.9545 | 79.5455 | laticauda_laticaudata | ENSLLTP00000023653 | 77.6596 | 74.4681 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 84.0909 | 80.6818 | notechis_scutatus | ENSNSUP00000014364 | 87.4016 | 83.8583 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 83.7121 | 79.9242 | pseudonaja_textilis | ENSPTXP00000004472 | 87.6984 | 83.7302 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 87.1212 | 84.4697 | gallus_gallus | ENSGALP00010043693 | 90.1961 | 87.451 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 54.9242 | 54.9242 | serinus_canaria | ENSSCAP00000015041 | 96.0265 | 96.0265 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 85.2273 | 83.7121 | parus_major | ENSPMJP00000020492 | 88.5827 | 87.0079 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 78.4091 | 74.6212 | anolis_carolinensis | ENSACAP00000010333 | 72.3776 | 68.8811 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 84.0909 | 80.6818 | naja_naja | ENSNNAP00000018639 | 87.4016 | 83.8583 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 84.4697 | 80.6818 | podarcis_muralis | ENSPMRP00000008590 | 67.7812 | 64.7416 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 51.1364 | 50.7576 | geospiza_fortis | ENSGFOP00000011739 | 93.75 | 93.0556 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 84.8485 | 83.7121 | taeniopygia_guttata | ENSTGUP00000003623 | 88.189 | 87.0079 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 82.9545 | 79.5455 | salvator_merianae | ENSSMRP00000030465 | 86.9048 | 83.3333 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 79.1667 | 75.7576 | oryzias_sinensis | ENSOSIP00000035720 | 77.9851 | 74.6269 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 48.4848 | 47.3485 | pelodiscus_sinensis | ENSPSIP00000019776 | 94.1176 | 91.9118 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 86.3636 | 83.7121 | gopherus_evgoodei | ENSGEVP00005021286 | 88.3721 | 85.6589 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 65.9091 | 56.8182 | seriola_dumerili | ENSSDUP00000005410 | 53.3742 | 46.0123 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 55.6818 | 43.9394 | cottoperca_gobio | ENSCGOP00000022985 | 55.6818 | 43.9394 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 86.7424 | 82.9545 | anser_brachyrhynchus | ENSABRP00000012496 | 69.1843 | 66.1631 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 65.9091 | 56.4394 | gasterosteus_aculeatus | ENSGACP00000024024 | 63.9706 | 54.7794 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 78.0303 | 73.8636 | gasterosteus_aculeatus | ENSGACP00000017621 | 79.845 | 75.5814 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 48.4848 | 41.6667 | cynoglossus_semilaevis | ENSCSEP00000024193 | 70.3297 | 60.4396 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 67.4242 | 59.4697 | myripristis_murdjan | ENSMMDP00005017811 | 72.0648 | 63.5628 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 79.9242 | 76.8939 | myripristis_murdjan | ENSMMDP00005007537 | 79.3233 | 76.3158 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 67.4242 | 59.0909 | sander_lucioperca | ENSSLUP00000028480 | 68.7259 | 60.2317 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 85.9848 | 84.0909 | strigops_habroptila | ENSSHBP00005012638 | 88.6719 | 86.7188 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 85.9848 | 83.3333 | terrapene_carolina_triunguis | ENSTMTP00000016492 | 87.9845 | 85.2713 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 67.0455 | 57.9545 | seriola_lalandi_dorsalis | ENSSLDP00000006761 | 67.5573 | 58.3969 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 79.9242 | 75.3788 | stegastes_partitus | ENSSPAP00000014818 | 75.089 | 70.8185 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 80.303 | 77.2727 | acanthochromis_polyacanthus | ENSAPOP00000015471 | 77.9412 | 75 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 86.3636 | 84.4697 | aquila_chrysaetos_chrysaetos | ENSACCP00020001191 | 88.716 | 86.7704 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 77.2727 | 73.8636 | takifugu_rubripes | ENSTRUP00000022188 | 76.4045 | 73.0337 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 64.3939 | 61.7424 | ficedula_albicollis | ENSFALP00000012445 | 76.2332 | 73.0942 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 78.7879 | 74.6212 | hippocampus_comes | ENSHCOP00000012072 | 78.1955 | 74.0602 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 86.7424 | 84.4697 | coturnix_japonica | ENSCJPP00005027265 | 89.8039 | 87.451 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 75.3788 | 70.4545 | scleropages_formosus | ENSSFOP00015005641 | 67.0034 | 62.6263 |
| ENSG00000183684 | homo_sapiens | ENSP00000421592 | 79.1667 | 75.3788 | leptobrachium_leishanense | ENSLLEP00000038050 | 72.8223 | 69.338 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000018 | involved_in regulation of DNA recombination | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0000346 | part_of transcription export complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0000398 | involved_in mRNA splicing, via spliceosome | 1 | IC | Homo_sapiens(9606) | Process |
| GO:0000781 | colocalizes_with chromosome, telomeric region | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0001649 | involved_in osteoblast differentiation | 1 | HDA | Homo_sapiens(9606) | Process |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0003729 | enables mRNA binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0006405 | involved_in RNA export from nucleus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006406 | involved_in mRNA export from nucleus | 2 | IBA,IMP | Homo_sapiens(9606) | Process |
| GO:0016020 | located_in membrane | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0016607 | located_in nuclear speck | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0031297 | involved_in replication fork processing | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032786 | involved_in positive regulation of DNA-templated transcription, elongation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0046784 | involved_in viral mRNA export from host cell nucleus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0062153 | enables C5-methylcytidine-containing RNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0070062 | located_in extracellular exosome | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0071013 | part_of catalytic step 2 spliceosome | 1 | IDA | Homo_sapiens(9606) | Component |