Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | CancerCHIKVSARS-COV-OC43FlavivirusesHIVRVSARS-COV-2SINVDengueZika |
| Drug | Drug |
| Main interacting RNAs | rRNA |
| Moonlighting functions | N.A. |
| Localizations | Stress granucleNucleoli rim |
| BulkPerturb-seq | DataSet_01_286 DataSet_01_366 |
Description
| Ensembl ID | ENSG00000181163 | Gene ID | 4869 | Accession | 7910 |
| Symbol | NPM1 | Alias | B23;NPM | Full Name | nucleophosmin 1 |
| Status | Confidence | Length | 24695 bases | Strand | Plus strand |
| Position | 5 : 171387116 - 171411810 | RNA binding domain | Nucleoplasmin | ||
| Summary | The protein encoded by this gene is involved in several cellular processes, including centrosome duplication, protein chaperoning, and cell proliferation. The encoded phosphoprotein shuttles between the nucleolus, nucleus, and cytoplasm, chaperoning ribosomal proteins and core histones from the nucleus to the cytoplasm. This protein is also known to sequester the tumor suppressor ARF in the nucleolus, protecting it from degradation until it is needed. Mutations in this gene are associated with acute myeloid leukemia. Dozens of pseudogenes of this gene have been identified. [provided by RefSeq, Aug 2017] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000503767 | Nucleoplasmin | PF03066 | 1.9e-40 | 1 |
| ENSP00000503150 | Nucleoplasmin | PF03066 | 2.3e-40 | 1 |
| ENSP00000503717 | Nucleoplasmin | PF03066 | 2.7e-40 | 1 |
| ENSP00000377408 | Nucleoplasmin | PF03066 | 5.3e-40 | 1 |
| ENSP00000503235 | Nucleoplasmin | PF03066 | 5.3e-40 | 1 |
| ENSP00000341168 | Nucleoplasmin | PF03066 | 5.7e-40 | 1 |
| ENSP00000296930 | Nucleoplasmin | PF03066 | 7.1e-40 | 1 |
| ENSP00000428755 | Nucleoplasmin | PF03066 | 7.1e-40 | 1 |
| ENSP00000504740 | Nucleoplasmin | PF03066 | 7.6e-40 | 1 |
| ENSP00000503283 | Nucleoplasmin | PF03066 | 8.8e-40 | 1 |
| ENSP00000504107 | Nucleoplasmin | PF03066 | 2.6e-22 | 1 |
| ENSP00000504016 | Nucleoplasmin | PF03066 | 3e-21 | 1 |
| ENSP00000503408 | Nucleoplasmin | PF03066 | 5.3e-21 | 1 |
| ENSP00000504308 | Nucleoplasmin | PF03066 | 2.7e-19 | 1 |
| ENSP00000429485 | Nucleoplasmin | PF03066 | 3.5e-19 | 1 |
| ENSP00000503781 | Nucleoplasmin | PF03066 | 3.5e-19 | 1 |
| ENSP00000428647 | Nucleoplasmin | PF03066 | 8.4e-10 | 1 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 29507312 | Analysis of the oligomeric states of nucleophosmin using size exclusion chromatography. | Gyosuke Sakashita | 2018-03-05 | Scientific reports |
| 21152184 | NPM1/B23: A Multifunctional Chaperone in Ribosome Biogenesis and Chromatin Remodeling. | S Mikael Lindström | 2011-01-01 | Biochemistry research international |
| 32937906 | MicroRNAs as Biomarkers and Therapeutic Targets in Inflammation- and Ischemia-Reperfusion-Related Acute Renal Injury. | Yueh-Lin Wu | 2020-09-14 | International journal of molecular sciences |
| 20615901 | Export of microRNAs and microRNA-protective protein by mammalian cells. | Kai Wang | 2010-11-01 | Nucleic acids research |
| 26950158 | Circulating microRNAs as Potential Biomarkers in Non-Alcoholic Fatty Liver Disease and Hepatocellular Carcinoma. | B Marta Afonso | 2016-03-03 | Journal of clinical medicine |
| 21425434 | Expression patterns of placental microRNAs. | Jean-Francois Mouillet | 2011-08-01 | Birth defects research. Part A, Clinical and molecular teratology |
| 22362753 | Function of homo- and hetero-oligomers of human nucleoplasmin/nucleophosmin family proteins NPM1, NPM2 and NPM3 during sperm chromatin remodeling. | Mitsuru Okuwaki | 2012-06-01 | Nucleic acids research |
| 21609964 | Characterization of extracellular circulating microRNA. | Andrey Turchinovich | 2011-09-01 | Nucleic acids research |
| 32451752 | MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. | Margherita Ratti | 2020-06-01 | Targeted oncology |
| 29462963 | Secreted and Tissue miRNAs as Diagnosis Biomarkers of Malignant Pleural Mesothelioma. | Vanessa Martínez-Rivera | 2018-02-17 | International journal of molecular sciences |
| 29312145 | MicroRNA Exocytosis by Vesicle Fusion in Neuroendocrine Cells. | Yongsoo Park | 2017-01-01 | Frontiers in endocrinology |
| 31015836 | Poly-ADP Ribosyl Polymerase 1 (PARP1) Regulates Influenza A Virus Polymerase. | Liset Westera | 2019-01-01 | Advances in virology |
| 22817753 | Extracellular small RNAs: what, where, why? | M Anna Hoy | 2012-08-01 | Biochemical Society transactions |
| 24273550 | Cardiovascular extracellular microRNAs: emerging diagnostic markers and mechanisms of cell-to-cell RNA communication. | Virginie Kinet | 2013-01-01 | Frontiers in genetics |
| 27446198 | Single Molecule Localization Microscopy of Mammalian Cell Nuclei on the Nanoscale. | Aleksander Szczurek | 2016-01-01 | Frontiers in genetics |
| 33502444 | High-fidelity reconstitution of stress granules and nucleoli in mammalian cellular lysate. | D Brian Freibaum | 2021-03-01 | The Journal of cell biology |
| 22546315 | Circulating microRNAs in cancer: origin, function and application. | Ruimin Ma | 2012-04-30 | Journal of experimental & clinical cancer research : CR |
| 27092141 | Is It worth Considering Circulating microRNAs in Multiple Sclerosis? | Ferdinand Jagot | 2016-01-01 | Frontiers in immunology |
| 23285058 | Elucidation of motifs in ribosomal protein S9 that mediate its nucleolar localization and binding to NPM1/nucleophosmin. | S Mikael Lindström | 2012-01-01 | PloS one |
| 24795753 | Expression of microRNAs in bovine and human pre-implantation embryo culture media. | Jenna Kropp | 2014-01-01 | Frontiers in genetics |
| 27669739 | The human nucleophosmin 1 mutation A inhibits myeloid differentiation of leukemia cells by modulating miR-10b. | Qin Zou | 2016-11-01 | Oncotarget |
| 33795683 | A KRAS-responsive long non-coding RNA controls microRNA processing. | Lei Shi | 2021-04-01 | Nature communications |
| 31447320 | Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. | A Marcelo Mori | 2019-10-01 | Cell metabolism |
| 30498217 | Compositional adaptability in NPM1-SURF6 scaffolding networks enabled by dynamic switching of phase separation mechanisms. | C Mylene Ferrolino | 2018-11-29 | Nature communications |
| 32093728 | Dipeptide repeat proteins inhibit homology-directed DNA double strand break repair in C9ORF72 ALS/FTD. | S Nadja Andrade | 2020-02-24 | Molecular neurodegeneration |
| 22028337 | Profiling of circulating microRNAs: from single biomarkers to re-wired networks. | Anna Zampetaki | 2012-03-15 | Cardiovascular research |
| 25655644 | Base excision DNA repair and cancer. | Gianluca Tell | 2015-01-20 | Oncotarget |
| 31952319 | Challenges in Using Circulating Micro-RNAs as Biomarkers for Cardiovascular Diseases. | Kyriacos Felekkis | 2020-01-15 | International journal of molecular sciences |
| 31067715 | Extracellular Vesicle Encapsulated MicroRNAs in Patients with Type 2 Diabetes Are Affected by Metformin Treatment. | Vikas Ghai | 2019-05-07 | Journal of clinical medicine |
| 33233600 | Extracellular MicroRNAs as Intercellular Mediators and Noninvasive Biomarkers of Cancer. | Blanca Ortiz-Quintero | 2020-11-20 | Cancers |
| 36142403 | MicroRNAs, Stem Cells in Bipolar Disorder, and Lithium Therapeutic Approach. | Donatella Coradduzza | 2022-09-10 | International journal of molecular sciences |
| 27596437 | Identification of a novel mechanism of blood-brain communication during peripheral inflammation via choroid plexus-derived extracellular vesicles. | Sriram Balusu | 2016-10-01 | EMBO molecular medicine |
| 24733970 | Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. | Yogesh Dwivedi | 2014-03-01 | Dialogues in clinical neuroscience |
| 26836305 | Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. | M Diana Mitrea | 2016-02-02 | eLife |
| 29516617 | Extracellular vesicle RNAs reflect placenta dysfunction and are a biomarker source for preterm labour. | Shannon Fallen | 2018-05-01 | Journal of cellular and molecular medicine |
| 20699270 | Critical lysine residues within the overlooked N-terminal domain of human APE1 regulate its biological functions. | Damiano Fantini | 2010-12-01 | Nucleic acids research |
| 32957662 | Circulating Cell-Free Nucleic Acids: Main Characteristics and Clinical Application. | Melinda Szilágyi | 2020-09-17 | International journal of molecular sciences |
| 29589547 | Implication of B23/NPM1 in Viral Infections, Potential Uses of B23/NPM1 Inhibitors as Antiviral Therapy. | Yadira Lobaina | 2019-01-01 | Infectious disorders drug targets |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| NPM1-204 | ENST00000517671 | 1338 | 294aa | ENSP00000428755 |
| NPM1-215 | ENST00000676613 | 2676 | 156aa | ENSP00000503767 |
| NPM1-227 | ENST00000678186 | 2674 | No protein | - |
| NPM1-228 | ENST00000678267 | 2797 | 99aa | ENSP00000504107 |
| NPM1-217 | ENST00000677297 | 883 | 123aa | ENSP00000504016 |
| NPM1-208 | ENST00000521672 | 1263 | 230aa | ENSP00000429485 |
| NPM1-232 | ENST00000679190 | 794 | 90aa | ENSP00000503408 |
| NPM1-229 | ENST00000678280 | 1913 | 259aa | ENSP00000503235 |
| NPM1-220 | ENST00000677467 | 2600 | No protein | - |
| NPM1-231 | ENST00000679006 | 1116 | No protein | - |
| NPM1-216 | ENST00000676625 | 3605 | No protein | - |
| NPM1-202 | ENST00000351986 | 1237 | 265aa | ENSP00000341168 |
| NPM1-214 | ENST00000676589 | 1338 | 323aa | ENSP00000503283 |
| NPM1-213 | ENST00000676504 | 1936 | No protein | - |
| NPM1-233 | ENST00000679233 | 1072 | 195aa | ENSP00000503717 |
| NPM1-219 | ENST00000677357 | 1346 | 305aa | ENSP00000504740 |
| NPM1-201 | ENST00000296930 | 1320 | 294aa | ENSP00000296930 |
| NPM1-203 | ENST00000393820 | 1598 | 259aa | ENSP00000377408 |
| NPM1-210 | ENST00000523339 | 268 | No protein | - |
| NPM1-209 | ENST00000521710 | 1021 | No protein | - |
| NPM1-205 | ENST00000518587 | 1382 | No protein | - |
| NPM1-230 | ENST00000678774 | 1289 | 175aa | ENSP00000503150 |
| NPM1-207 | ENST00000521260 | 1571 | No protein | - |
| NPM1-221 | ENST00000677600 | 2506 | No protein | - |
| NPM1-224 | ENST00000677741 | 2459 | No protein | - |
| NPM1-211 | ENST00000523622 | 247 | 59aa | ENSP00000428647 |
| NPM1-218 | ENST00000677325 | 2258 | 230aa | ENSP00000503781 |
| NPM1-222 | ENST00000677672 | 2627 | No protein | - |
| NPM1-226 | ENST00000677907 | 1335 | 201aa | ENSP00000504308 |
| NPM1-225 | ENST00000677904 | 1469 | No protein | - |
| NPM1-223 | ENST00000677682 | 2534 | No protein | - |
| NPM1-206 | ENST00000519955 | 571 | No protein | - |
| NPM1-212 | ENST00000524204 | 640 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 31172 | Erythrocyte Indices | 8.3100000E-005 | - |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
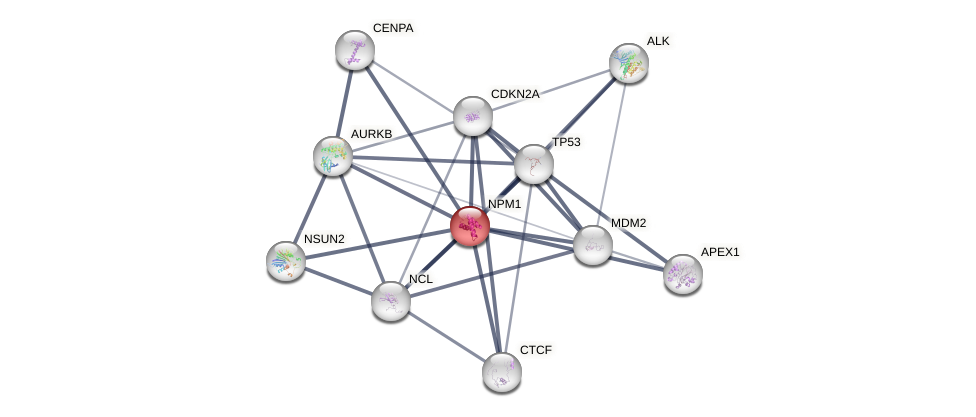
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000181163 | homo_sapiens | ENSP00000359128 | 51.6854 | 34.8315 | homo_sapiens | ENSP00000296930 | 31.2925 | 21.0884 |
| ENSG00000181163 | homo_sapiens | ENSP00000427741 | 50.9346 | 30.8411 | homo_sapiens | ENSP00000296930 | 37.0748 | 22.449 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 100 | 99.6599 | nomascus_leucogenys | ENSNLEP00000000400 | 100 | 99.6599 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 99.6599 | 99.6599 | pan_paniscus | ENSPPAP00000038345 | 99.6599 | 99.6599 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 90.1361 | 89.7959 | pongo_abelii | ENSPPYP00000017949 | 99.6241 | 99.2481 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 100 | 100 | pan_troglodytes | ENSPTRP00000068344 | 100 | 100 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 100 | 100 | gorilla_gorilla | ENSGGOP00000037355 | 100 | 100 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 86.0544 | 72.449 | ornithorhynchus_anatinus | ENSOANP00000012413 | 84.6154 | 71.2375 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 96.5986 | 88.0952 | sarcophilus_harrisii | ENSSHAP00000027342 | 96.9283 | 88.3959 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 99.3197 | 98.2993 | callithrix_jacchus | ENSCJAP00000035920 | 98.6487 | 97.6351 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 95.5782 | 94.5578 | callithrix_jacchus | ENSCJAP00000054920 | 97.9094 | 96.8641 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 100 | 99.6599 | cercocebus_atys | ENSCATP00000003396 | 100 | 99.6599 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 90.1361 | 89.7959 | macaca_fascicularis | ENSMFAP00000023936 | 100 | 99.6226 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 100 | 99.6599 | macaca_mulatta | ENSMMUP00000020737 | 100 | 99.6599 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 100 | 99.3197 | macaca_nemestrina | ENSMNEP00000031336 | 100 | 99.3197 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 90.1361 | 89.7959 | papio_anubis | ENSPANP00000022161 | 100 | 99.6226 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 100 | 99.6599 | mandrillus_leucophaeus | ENSMLEP00000013536 | 100 | 99.6599 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 73.4694 | 69.3878 | mus_spretus | MGP_SPRETEiJ_P0090351 | 94.7368 | 89.4737 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 97.2789 | 94.898 | mus_spretus | MGP_SPRETEiJ_P0027462 | 97.9452 | 95.5479 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 98.2993 | 95.2381 | loxodonta_africana | ENSLAFP00000006449 | 98.2993 | 95.2381 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 75.8503 | 73.8095 | loxodonta_africana | ENSLAFP00000025474 | 94.4915 | 91.9492 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 59.5238 | 54.0816 | loxodonta_africana | ENSLAFP00000028188 | 83.7321 | 76.0766 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 100 | 100 | ailuropoda_melanoleuca | ENSAMEP00000023639 | 100 | 100 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 99.3197 | 97.619 | oryctolagus_cuniculus | ENSOCUP00000009569 | 99.3197 | 97.619 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 76.1905 | 72.7891 | oryctolagus_cuniculus | ENSOCUP00000020753 | 92.9461 | 88.7967 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 99.6599 | 97.619 | oryctolagus_cuniculus | ENSOCUP00000021783 | 93.6102 | 91.6933 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 96.9388 | 94.898 | oryctolagus_cuniculus | ENSOCUP00000023192 | 97.6027 | 95.5479 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 91.4966 | 80.9524 | monodelphis_domestica | ENSMODP00000017801 | 92.4399 | 81.7869 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 91.4966 | 81.6327 | monodelphis_domestica | ENSMODP00000008071 | 97.1119 | 86.6426 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 71.4286 | 70.068 | microcebus_murinus | ENSMICP00000026690 | 96.7742 | 94.9309 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 88.4354 | 86.0544 | microcebus_murinus | ENSMICP00000051154 | 98.1132 | 95.4717 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 99.3197 | 97.9592 | microcebus_murinus | ENSMICP00000048331 | 98.983 | 97.6271 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 65.6463 | 62.585 | octodon_degus | ENSODEP00000002466 | 96.5 | 92 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 78.2313 | 77.551 | ovis_aries_rambouillet | ENSOARP00020033944 | 97.4576 | 96.6102 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 81.9728 | 79.2517 | ochotona_princeps | ENSOPRP00000006764 | 87.9562 | 85.0365 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 97.619 | 94.2177 | mus_caroli | MGP_CAROLIEiJ_P0026112 | 98.2877 | 94.863 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 97.619 | 94.2177 | mus_musculus | ENSMUSP00000075067 | 98.2877 | 94.863 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 86.7347 | 82.6531 | dipodomys_ordii | ENSDORP00000027268 | 96.5909 | 92.0455 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 92.1769 | 88.4354 | dipodomys_ordii | ENSDORP00000022161 | 99.2674 | 95.2381 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 67.3469 | 62.585 | dipodomys_ordii | ENSDORP00000022299 | 94.7368 | 88.0383 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 100 | 99.6599 | chlorocebus_sabaeus | ENSCSAP00000005927 | 100 | 99.6599 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 97.619 | 94.2177 | rattus_norvegicus | ENSRNOP00000006591 | 81.7664 | 78.9174 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 78.2313 | 74.4898 | rattus_norvegicus | ENSRNOP00000058202 | 92.7419 | 88.3064 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 82.3129 | 79.2517 | rattus_norvegicus | ENSRNOP00000095107 | 95.2756 | 91.7323 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 48.9796 | 44.5578 | rattus_norvegicus | ENSRNOP00000074426 | 87.2727 | 79.3939 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 83.6735 | 77.551 | rattus_norvegicus | ENSRNOP00000088215 | 89.781 | 83.2117 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 94.898 | 91.4966 | rattus_norvegicus | ENSRNOP00000025575 | 96.5398 | 93.0796 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 93.8775 | 88.7755 | microtus_ochrogaster | ENSMOCP00000005839 | 94.198 | 89.0785 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 98.2993 | 95.2381 | microtus_ochrogaster | ENSMOCP00000001917 | 98.9726 | 95.8904 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 75.1701 | 71.4286 | microtus_ochrogaster | ENSMOCP00000002083 | 96.087 | 91.3043 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 76.5306 | 70.7483 | microtus_ochrogaster | ENSMOCP00000022394 | 96.1538 | 88.8889 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 81.6327 | 75.8503 | microtus_ochrogaster | ENSMOCP00000026646 | 91.6031 | 85.1145 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 52.381 | 48.6395 | peromyscus_maniculatus_bairdii | ENSPEMP00000005980 | 70.6422 | 65.5963 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 81.9728 | 78.5714 | peromyscus_maniculatus_bairdii | ENSPEMP00000022482 | 96.0159 | 92.0319 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 85.034 | 81.6327 | peromyscus_maniculatus_bairdii | ENSPEMP00000033287 | 99.2064 | 95.2381 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 98.2993 | 95.2381 | peromyscus_maniculatus_bairdii | ENSPEMP00000008365 | 98.9726 | 95.8904 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 80.9524 | 76.5306 | mus_spicilegus | ENSMSIP00000017786 | 96.3563 | 91.0931 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 95.5782 | 92.517 | mus_spicilegus | ENSMSIP00000014008 | 97.9094 | 94.7735 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 83.3333 | 80.2721 | mus_spicilegus | ENSMSIP00000031792 | 96.4567 | 92.9134 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 80.9524 | 77.2109 | mus_spicilegus | ENSMSIP00000001149 | 90.4943 | 86.3118 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 97.619 | 94.2177 | mus_spicilegus | ENSMSIP00000027933 | 98.2877 | 94.863 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 66.3265 | 63.6054 | cricetulus_griseus_chok1gshd | ENSCGRP00001006263 | 94.2029 | 90.3382 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 76.5306 | 69.7279 | cricetulus_griseus_chok1gshd | ENSCGRP00001011672 | 88.9328 | 81.0277 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 86.0544 | 80.9524 | cricetulus_griseus_chok1gshd | ENSCGRP00001009786 | 96.1977 | 90.4943 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 97.9592 | 94.5578 | cricetulus_griseus_chok1gshd | ENSCGRP00001001990 | 98.6301 | 95.2055 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 79.932 | 74.8299 | cricetulus_griseus_chok1gshd | ENSCGRP00001006798 | 91.4397 | 85.6031 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 73.4694 | 56.1224 | vombatus_ursinus | ENSVURP00010004192 | 80.2974 | 61.3383 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 96.5986 | 87.7551 | vombatus_ursinus | ENSVURP00010001733 | 96.9283 | 88.0546 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 100 | 99.6599 | rhinopithecus_bieti | ENSRBIP00000038012 | 100 | 99.6599 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 96.5986 | 87.7551 | phascolarctos_cinereus | ENSPCIP00000017850 | 96.9283 | 88.0546 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 77.551 | 68.0272 | phascolarctos_cinereus | ENSPCIP00000039589 | 90.1186 | 79.0514 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 49.3197 | 45.9184 | phascolarctos_cinereus | ENSPCIP00000016942 | 96.0265 | 89.404 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 86.7347 | 83.6735 | procavia_capensis | ENSPCAP00000006117 | 87.3288 | 84.2466 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 99.3197 | 98.2993 | catagonus_wagneri | ENSCWAP00000001295 | 99.3197 | 98.2993 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 98.2993 | 96.5986 | urocitellus_parryii | ENSUPAP00010028066 | 98.9726 | 97.2603 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 100 | 99.6599 | rhinopithecus_roxellana | ENSRROP00000044358 | 100 | 99.6599 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 88.4354 | 84.0136 | mus_pahari | MGP_PahariEiJ_P0032884 | 94.8905 | 90.146 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 85.3742 | 80.9524 | propithecus_coquereli | ENSPCOP00000022967 | 90.6137 | 85.9206 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 29.2517 | 18.7075 | drosophila_melanogaster | FBpp0084918 | 56.5789 | 36.1842 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 27.8912 | 17.0068 | drosophila_melanogaster | FBpp0084971 | 52.5641 | 32.0513 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 57.483 | 36.3946 | ciona_intestinalis | ENSCINP00000033019 | 51.0574 | 32.3263 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 53.0612 | 35.7143 | eptatretus_burgeri | ENSEBUP00000020602 | 40.3101 | 27.1318 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 56.4626 | 41.1565 | lepisosteus_oculatus | ENSLOCP00000011829 | 62.6415 | 45.6604 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 65.6463 | 48.2993 | danio_rerio | ENSDARP00000117965 | 69.1756 | 50.8961 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 64.2857 | 48.2993 | carassius_auratus | ENSCARP00000084572 | 61.5635 | 46.2541 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 63.6054 | 44.2177 | carassius_auratus | ENSCARP00000118529 | 65.8451 | 45.7746 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 65.9864 | 45.9184 | carassius_auratus | ENSCARP00000030213 | 66.6667 | 46.3918 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 60.8844 | 44.5578 | astyanax_mexicanus | ENSAMXP00000010479 | 69.112 | 50.5792 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 66.3265 | 47.619 | ictalurus_punctatus | ENSIPUP00000028757 | 66.7808 | 47.9452 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 65.6463 | 45.5782 | esox_lucius | ENSELUP00000013626 | 65.8703 | 45.7338 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 67.6871 | 49.6599 | electrophorus_electricus | ENSEEEP00000002287 | 65.6766 | 48.1848 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 52.7211 | 38.4354 | oncorhynchus_kisutch | ENSOKIP00005115488 | 58.0524 | 42.3221 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 60.8844 | 43.5374 | oncorhynchus_kisutch | ENSOKIP00005076994 | 60.473 | 43.2432 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 60.5442 | 44.5578 | oncorhynchus_mykiss | ENSOMYP00000105556 | 63.3452 | 46.6192 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 60.5442 | 43.1973 | oncorhynchus_mykiss | ENSOMYP00000122565 | 58.9404 | 42.053 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 63.2653 | 44.898 | salmo_salar | ENSSSAP00000180712 | 56.535 | 40.1216 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 57.483 | 42.517 | salmo_salar | ENSSSAP00000065751 | 62.1324 | 45.9559 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 64.6258 | 45.9184 | salmo_trutta | ENSSTUP00000046680 | 63.7584 | 45.302 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 60.5442 | 44.2177 | salmo_trutta | ENSSTUP00000020447 | 63.7993 | 46.595 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 62.2449 | 44.2177 | gadus_morhua | ENSGMOP00000057733 | 63.3218 | 44.9827 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 64.2857 | 45.9184 | fundulus_heteroclitus | ENSFHEP00000011113 | 63 | 45 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 65.9864 | 48.6395 | poecilia_reticulata | ENSPREP00000002220 | 64.8829 | 47.8261 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 63.9456 | 46.5986 | xiphophorus_maculatus | ENSXMAP00000000395 | 62.0462 | 45.2145 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 64.6258 | 45.9184 | oryzias_latipes | ENSORLP00000012763 | 65.0685 | 46.2329 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 32.3129 | 25.1701 | cyclopterus_lumpus | ENSCLMP00005044404 | 53.9773 | 42.0455 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 55.102 | 39.7959 | oreochromis_niloticus | ENSONIP00000071006 | 56.8421 | 41.0526 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 67.6871 | 48.2993 | haplochromis_burtoni | ENSHBUP00000020177 | 63.1746 | 45.0794 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 54.4218 | 40.8163 | astatotilapia_calliptera | ENSACLP00000012279 | 59.2593 | 44.4444 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 68.3673 | 50.3401 | sparus_aurata | ENSSAUP00010005086 | 67.2241 | 49.4983 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 55.102 | 38.0952 | lates_calcarifer | ENSLCAP00010045292 | 57.6512 | 39.8577 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 63.6054 | 44.5578 | neolamprologus_brichardi | ENSNBRP00000017062 | 60.9121 | 42.671 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 63.6054 | 43.8776 | kryptolebias_marmoratus | ENSKMAP00000014280 | 64.2612 | 44.3299 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 61.9048 | 44.2177 | oryzias_javanicus | ENSOJAP00000039491 | 57.9618 | 41.4013 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 59.8639 | 43.8776 | amphilophus_citrinellus | ENSACIP00000018212 | 64.7059 | 47.4265 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 67.3469 | 48.9796 | dicentrarchus_labrax | ENSDLAP00005053491 | 66.2207 | 48.1605 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 66.3265 | 48.2993 | amphiprion_percula | ENSAPEP00000012037 | 65.4362 | 47.651 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 51.3605 | 35.7143 | oryzias_sinensis | ENSOSIP00000028107 | 64.8069 | 45.0644 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 67.3469 | 45.5782 | seriola_dumerili | ENSSDUP00000021441 | 66.443 | 44.9664 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 62.585 | 44.898 | cottoperca_gobio | ENSCGOP00000056409 | 63.8889 | 45.8333 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 62.2449 | 43.5374 | gasterosteus_aculeatus | ENSGACP00000026847 | 59.8039 | 41.8301 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 53.7415 | 37.415 | cynoglossus_semilaevis | ENSCSEP00000017415 | 65.0206 | 45.2675 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 48.9796 | 34.3537 | cynoglossus_semilaevis | ENSCSEP00000031793 | 38.0952 | 26.7196 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 65.9864 | 47.9592 | poecilia_formosa | ENSPFOP00000019288 | 64.8829 | 47.1572 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 47.2789 | 30.6122 | myripristis_murdjan | ENSMMDP00005054714 | 48.4321 | 31.3589 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 63.9456 | 46.9388 | sander_lucioperca | ENSSLUP00000012412 | 65.0519 | 47.7509 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 52.381 | 39.7959 | amphiprion_ocellaris | ENSAOCP00000001882 | 66.6667 | 50.6493 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 60.5442 | 44.5578 | hucho_hucho | ENSHHUP00000043335 | 63.5714 | 46.7857 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 64.966 | 46.5986 | hucho_hucho | ENSHHUP00000073652 | 63.4552 | 45.5149 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 57.1429 | 42.1769 | betta_splendens | ENSBSLP00000055557 | 59.1549 | 43.662 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 64.2857 | 45.2381 | pygocentrus_nattereri | ENSPNAP00000036221 | 63.6364 | 44.7811 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 35.3741 | 26.1905 | anabas_testudineus | ENSATEP00000068256 | 56.8306 | 42.0765 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 59.1837 | 36.7347 | ciona_savignyi | ENSCSAVP00000019066 | 51.3274 | 31.8584 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 67.3469 | 46.5986 | seriola_lalandi_dorsalis | ENSSLDP00000022864 | 66.6667 | 46.1279 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 65.6463 | 46.9388 | labrus_bergylta | ENSLBEP00000011405 | 64.7651 | 46.3087 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 67.0068 | 48.2993 | pundamilia_nyererei | ENSPNYP00000025619 | 63.5484 | 45.8064 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 67.0068 | 50.3401 | mastacembelus_armatus | ENSMAMP00000013775 | 55.1821 | 41.4566 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 55.4422 | 41.8367 | stegastes_partitus | ENSSPAP00000028784 | 65.7258 | 49.5968 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 59.1837 | 43.8776 | oncorhynchus_tshawytscha | ENSOTSP00005005294 | 62.5899 | 46.4029 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 62.9252 | 44.898 | oncorhynchus_tshawytscha | ENSOTSP00005010212 | 62.9252 | 44.898 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 67.0068 | 49.3197 | acanthochromis_polyacanthus | ENSAPOP00000001512 | 65.8863 | 48.495 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 62.585 | 46.2585 | nothobranchius_furzeri | ENSNFUP00015054039 | 64.1115 | 47.3868 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 63.9456 | 45.5782 | cyprinodon_variegatus | ENSCVAP00000024705 | 63.5135 | 45.2703 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 49.3197 | 32.3129 | denticeps_clupeoides | ENSDCDP00000003717 | 58.9431 | 38.6179 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 35.034 | 24.4898 | denticeps_clupeoides | ENSDCDP00000003765 | 56.5934 | 39.5604 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 67.3469 | 46.2585 | denticeps_clupeoides | ENSDCDP00000003654 | 68.5121 | 47.0588 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 41.1565 | 26.5306 | denticeps_clupeoides | ENSDCDP00000003769 | 64.3617 | 41.4894 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 31.9728 | 25.5102 | larimichthys_crocea | ENSLCRP00005055915 | 61.039 | 48.7013 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 65.9864 | 45.5782 | takifugu_rubripes | ENSTRUP00000055343 | 61.7834 | 42.6752 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 58.8435 | 42.517 | hippocampus_comes | ENSHCOP00000000277 | 65.283 | 47.1698 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 67.0068 | 49.3197 | cyprinus_carpio_carpio | ENSCCRP00000106894 | 63.5484 | 46.7742 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 58.1633 | 39.7959 | cyprinus_carpio_carpio | ENSCCRP00000172976 | 63.3333 | 43.3333 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 64.966 | 48.2993 | poecilia_latipinna | ENSPLAP00000000286 | 64.094 | 47.651 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 66.6667 | 47.619 | sinocyclocheilus_grahami | ENSSGRP00000095361 | 67.8201 | 48.4429 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 67.0068 | 46.9388 | sinocyclocheilus_grahami | ENSSGRP00000112630 | 59.8784 | 41.9453 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 66.6667 | 47.2789 | maylandia_zebra | ENSMZEP00005018273 | 62.6198 | 44.4089 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 62.585 | 43.5374 | tetraodon_nigroviridis | ENSTNIP00000013697 | 63.2302 | 43.9863 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 64.6258 | 48.2993 | paramormyrops_kingsleyae | ENSPKIP00000008462 | 63.1229 | 47.1761 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 63.6054 | 49.3197 | scleropages_formosus | ENSSFOP00015010908 | 65.8451 | 51.0563 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 59.1837 | 45.9184 | scophthalmus_maximus | ENSSMAP00000031177 | 56.6775 | 43.9739 |
| ENSG00000181163 | homo_sapiens | ENSP00000296930 | 64.2857 | 45.2381 | oryzias_melastigma | ENSOMEP00000006043 | 64.5051 | 45.3925 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000055 | involved_in ribosomal large subunit export from nucleus | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0000056 | involved_in ribosomal small subunit export from nucleus | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0001046 | enables core promoter sequence-specific DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0001558 | involved_in regulation of cell growth | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001652 | located_in granular component | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0003682 | enables chromatin binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0003713 | enables transcription coactivator activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 4 | HDA,IBA,IDA,IPI | Homo_sapiens(9606) | Function |
| GO:0004860 | enables protein kinase inhibitor activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 2 | HDA,IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | is_active_in nucleoplasm | 3 | IBA,IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005730 | is_active_in nucleolus | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005813 | is_active_in centrosome | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005925 | located_in focal adhesion | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0006281 | involved_in DNA repair | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006334 | involved_in nucleosome assembly | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006338 | involved_in chromatin remodeling | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0006407 | involved_in rRNA export from nucleus | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0006884 | involved_in cell volume homeostasis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006886 | involved_in intracellular protein transport | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006913 | involved_in nucleocytoplasmic transport | 2 | IDA,TAS | Homo_sapiens(9606) | Process |
| GO:0007098 | involved_in centrosome cycle | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0007165 | involved_in signal transduction | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0008104 | involved_in protein localization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0008284 | involved_in positive regulation of cell population proliferation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0008285 | involved_in negative regulation of cell population proliferation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0010824 | involved_in regulation of centrosome duplication | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0010825 | involved_in positive regulation of centrosome duplication | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010826 | involved_in negative regulation of centrosome duplication | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0015934 | part_of large ribosomal subunit | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0015935 | part_of small ribosomal subunit | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0016020 | located_in membrane | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0016607 | located_in nuclear speck | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0019843 | enables rRNA binding | 1 | EXP | Homo_sapiens(9606) | Function |
| GO:0019901 | enables protein kinase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0030957 | enables Tat protein binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0031398 | involved_in positive regulation of protein ubiquitination | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0031616 | located_in spindle pole centrosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0032071 | involved_in regulation of endodeoxyribonuclease activity | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032991 | part_of protein-containing complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0032993 | part_of protein-DNA complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0034644 | involved_in cellular response to UV | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0042255 | involved_in ribosome assembly | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0042273 | involved_in ribosomal large subunit biogenesis | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0042274 | involved_in ribosomal small subunit biogenesis | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0042393 | enables histone binding | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0042803 | enables protein homodimerization activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0043023 | enables ribosomal large subunit binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0043024 | enables ribosomal small subunit binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0043066 | involved_in negative regulation of apoptotic process | 2 | IDA,NAS | Homo_sapiens(9606) | Process |
| GO:0043516 | involved_in regulation of DNA damage response, signal transduction by p53 class mediator | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0044387 | involved_in negative regulation of protein kinase activity by regulation of protein phosphorylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045727 | involved_in positive regulation of translation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045860 | involved_in positive regulation of protein kinase activity | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045893 | involved_in positive regulation of DNA-templated transcription | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 3 | IBA,IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0046599 | involved_in regulation of centriole replication | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0048025 | involved_in negative regulation of mRNA splicing, via spliceosome | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0050821 | involved_in protein stabilization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051059 | enables NF-kappaB binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0051082 | enables unfolded protein binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0051092 | involved_in positive regulation of NF-kappaB transcription factor activity | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0060699 | involved_in regulation of endoribonuclease activity | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0060735 | involved_in regulation of eIF2 alpha phosphorylation by dsRNA | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0090398 | involved_in cellular senescence | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0140297 | enables DNA-binding transcription factor binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0140693 | enables molecular condensate scaffold activity | 2 | EXP,IDA | Homo_sapiens(9606) | Function |
| GO:1902629 | involved_in regulation of mRNA stability involved in cellular response to UV | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1902751 | involved_in positive regulation of cell cycle G2/M phase transition | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1904751 | involved_in positive regulation of protein localization to nucleolus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1990904 | part_of ribonucleoprotein complex | 2 | IBA,IDA | Homo_sapiens(9606) | Component |