Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | N.A. |
| Drug | N.A. |
| Main interacting RNAs | snoRNA |
| Moonlighting functions | N.A. |
| Localizations | Nucleoli |
| BulkPerturb-seq | N.A. |
Description
| Ensembl ID | ENSG00000177971 | Gene ID | 55272 | Accession | 14497 |
| Symbol | IMP3 | Alias | BRMS2;MRPS4;C15orf12 | Full Name | IMP U3 small nucleolar ribonucleoprotein 3 |
| Status | Confidence | Length | 9622 bases | Strand | Minus strand |
| Position | 15 : 75639085 - 75648706 | RNA binding domain | Ribosomal_S4 , S4 | ||
| Summary | This gene encodes the human homolog of the yeast Imp3 protein. The protein localizes to the nucleoli and interacts with the U3 snoRNP complex. The protein contains an S4 domain. [provided by RefSeq, Jul 2008] | ||||
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 34703650 | Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. | Ting Yu | 2021-12-03 | Molecular therapy. Nucleic acids |
| 25295085 | Expression of insulin-like growth factor-II mRNA-binding protein-3 as a marker for predicting clinical outcome in patients with esophageal squamous cell carcinoma. | Akihiro Takata | 2014-11-01 | Oncology letters |
| 32616503 | Y Chromosome LncRNA Are Involved in Radiation Response of Male Non-Small Cell Lung Cancer Cells. | Tayvia Brownmiller | 2020-10-01 | Cancer research |
| 28828031 | A diagnostically difficult case of a cellular pleural fluid: Morphology, immunohistochemistry, and fluorescence in situ hybridization study. | Minhua Wang | 2017-01-01 | CytoJournal |
| 32293476 | IGF2BP3 (IMP3) expression in angiosarcoma, epithelioid hemangioendothelioma, and benign vascular lesions. | Misuzu Okabayshi | 2020-03-23 | Diagnostic pathology |
| 21613208 | Insulin growth factor-2 binding protein 3 (IGF2BP3) is a glioblastoma-specific marker that activates phosphatidylinositol 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) pathways by modulating IGF-2. | Ramaswamy Suvasini | 2011-07-22 | The Journal of biological chemistry |
| 31118463 | Combinatorial recognition of clustered RNA elements by the multidomain RNA-binding protein IMP3. | Tim Schneider | 2019-05-22 | Nature communications |
| 21809995 | The oncofetal protein IMP3: a novel molecular marker to predict aggressive meningioma. | Suyang Hao | 2011-08-01 | Archives of pathology & laboratory medicine |
| 26123742 | PIpelle Prospective ENDOmetrial carcinoma (PIPENDO) study, pre-operative recognition of high risk endometrial carcinoma: a multicentre prospective cohort study. | M C Nicole Visser | 2015-06-30 | BMC cancer |
| 26796627 | Insulin-Like Growth Factor II mRNA-Binding Protein 3 Expression Correlates with Poor Prognosis in Acral Lentiginous Melanoma. | Yi-Shuan Sheen | 2016-01-01 | PloS one |
| 28654463 | Fatty Acid Synthase and Acetyl-CoA Carboxylase Are Expressed in Nodal Metastatic Melanoma But Not in Benign Intracapsular Nodal Nevi. | Jad Saab | 2018-04-01 | The American Journal of dermatopathology |
| 25982283 | IMP3 promotes stem-like properties in triple-negative breast cancer by regulating SLUG. | S Samanta | 2016-03-03 | Oncogene |
| 24411949 | Insulin-like growth factor II messenger RNA-binding protein 3 expression in gastrointestinal mesenchymal tumors. | Hidetaka Yamamoto | 2014-03-01 | Human pathology |
| 32365712 | High Mobility Group AT-Hook 2 (HMGA2) Oncogenicity in Mesenchymal and Epithelial Neoplasia. | Uchenna Unachukwu | 2020-04-29 | International journal of molecular sciences |
| 36819630 | The Expression of Insulin-Like Growth Factor II Messenger RNA-Binding Protein 3 Upregulated in Intradural Extramedullary Schwannomas. | Hirofumi Bekki | 2023-01-27 | Spine surgery and related research |
| 27510448 | CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. | Tim Schneider | 2016-08-11 | Scientific reports |
| 18204432 | IMP-3 is a novel progression marker in malignant melanoma. | G Jennifer Pryor | 2008-04-01 | Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc |
| 26982091 | The RNA binding protein IMP3 facilitates tumor immune escape by downregulating the stress-induced ligands ULPB2 and MICB. | Dominik Schmiedel | 2016-03-16 | eLife |
| 16814207 | Analysis of RNA-binding protein IMP3 to predict metastasis and prognosis of renal-cell carcinoma: a retrospective study. | Zhong Jiang | 2006-07-01 | The Lancet. Oncology |
| 15753088 | The RNA-binding protein IMP-3 is a translational activator of insulin-like growth factor II leader-3 mRNA during proliferation of human K562 leukemia cells. | Baisong Liao | 2005-05-06 | The Journal of biological chemistry |
| 34503117 | IMP3 Protein Overexpression Is Linked to Unfavorable Outcome in Laryngeal Squamous Cell Carcinoma. | Diana Maržić | 2021-08-26 | Cancers |
| 25759769 | Imp3 expression in benign and malignant thyroid tumors and hyperplastic nodules. | Sezer Kulaçoğlu | 2015-01-01 | Balkan medical journal |
| 20060157 | IMP3/L523S, a novel immunocytochemical marker that distinguishes benign and malignant cells: the expression profiles of IMP3/L523S in effusion cytology. | Katsuhide Ikeda | 2010-05-01 | Human pathology |
| 30135093 | Structural basis of IMP3 RRM12 recognition of RNA. | Min Jia | 2018-12-01 | RNA (New York, N.Y.) |
| 29847788 | IMP3 Stabilization of WNT5B mRNA Facilitates TAZ Activation in Breast Cancer. | Sanjoy Samanta | 2018-05-29 | Cell reports |
| 23465280 | Expression of the oncofetal protein IGF2BP3 in endometrial clear cell carcinoma: assessment of frequency and significance. | Oluwole Fadare | 2013-08-01 | Human pathology |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| IMP3-201 | ENST00000314852 | 2028 | 184aa | ENSP00000326981 |
| IMP3-203 | ENST00000565349 | 557 | No protein | - |
| IMP3-202 | ENST00000403490 | 1132 | 184aa | ENSP00000385217 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
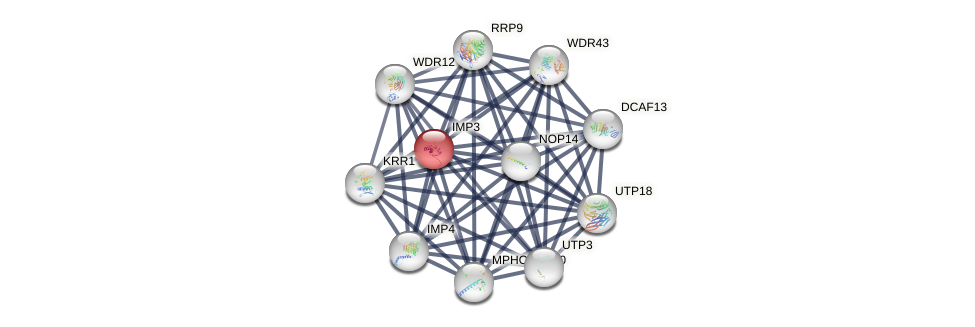
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000177971 | ||||||||
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 100 | 100 | pan_paniscus | ENSPPAP00000002045 | 100 | 100 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 100 | 99.4565 | pongo_abelii | ENSPPYP00000007559 | 100 | 99.4565 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 100 | 100 | pan_troglodytes | ENSPTRP00000077106 | 100 | 100 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 100 | 100 | gorilla_gorilla | ENSGGOP00000044840 | 100 | 100 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 90.2174 | 79.8913 | ornithorhynchus_anatinus | ENSOANP00000033842 | 90.2174 | 79.8913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 92.9348 | 85.3261 | sarcophilus_harrisii | ENSSHAP00000007136 | 73.7069 | 67.6724 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 97.8261 | choloepus_hoffmanni | ENSCHOP00000010641 | 98.913 | 97.8261 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 97.8261 | dasypus_novemcinctus | ENSDNOP00000012245 | 80.6167 | 79.2952 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.3696 | 95.6522 | echinops_telfairi | ENSETEP00000003501 | 98.3696 | 95.6522 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.913 | callithrix_jacchus | ENSCJAP00000055068 | 99.4565 | 98.913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 100 | 100 | cercocebus_atys | ENSCATP00000004244 | 100 | 100 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 100 | 100 | macaca_fascicularis | ENSMFAP00000010654 | 100 | 100 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 100 | 100 | macaca_mulatta | ENSMMUP00000021183 | 100 | 100 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 100 | 100 | macaca_nemestrina | ENSMNEP00000005835 | 100 | 100 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 100 | 100 | papio_anubis | ENSPANP00000009940 | 100 | 100 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 97.8261 | 95.6522 | vulpes_vulpes | ENSVVUP00000026659 | 97.8261 | 95.6522 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 96.1957 | mus_spretus | MGP_SPRETEiJ_P0091602 | 98.913 | 96.1957 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.913 | equus_asinus | ENSEASP00005059555 | 99.4565 | 98.913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | capra_hircus | ENSCHIP00000002648 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 97.8261 | 97.2826 | loxodonta_africana | ENSLAFP00000003692 | 97.8261 | 97.2826 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 99.4565 | panthera_leo | ENSPLOP00000002125 | 99.4565 | 99.4565 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | ailuropoda_melanoleuca | ENSAMEP00000020312 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | bos_indicus_hybrid | ENSBIXP00005005289 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 98.913 | oryctolagus_cuniculus | ENSOCUP00000004048 | 46.7866 | 46.7866 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 99.4565 | equus_caballus | ENSECAP00000025672 | 99.4565 | 99.4565 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 76.087 | 73.913 | canis_lupus_familiaris | ENSCAFP00845010825 | 80.9249 | 78.6127 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 97.8261 | 96.1957 | canis_lupus_familiaris | ENSCAFP00845028470 | 97.8261 | 96.1957 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 99.4565 | panthera_pardus | ENSPPRP00000027333 | 99.4565 | 99.4565 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 98.3696 | marmota_marmota_marmota | ENSMMMP00000002333 | 98.913 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | balaenoptera_musculus | ENSBMSP00010021210 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 97.8261 | cavia_porcellus | ENSCPOP00000020803 | 99.4565 | 97.8261 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.913 | ursus_americanus | ENSUAMP00000029496 | 99.4565 | 98.913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 92.9348 | 85.3261 | monodelphis_domestica | ENSMODP00000037347 | 92.9348 | 85.3261 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | bison_bison_bison | ENSBBBP00000017702 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | monodon_monoceros | ENSMMNP00015014203 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 98.913 | sus_scrofa | ENSSSCP00000002051 | 98.913 | 98.913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 98.3696 | microcebus_murinus | ENSMICP00000048440 | 98.913 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 97.8261 | octodon_degus | ENSODEP00000026729 | 99.4565 | 97.8261 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 96.1957 | 94.0217 | ovis_aries_rambouillet | ENSOARP00020032656 | 96.1957 | 94.0217 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | ovis_aries_rambouillet | ENSOARP00020028178 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.913 | ursus_maritimus | ENSUMAP00000004630 | 99.4565 | 98.913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 97.2826 | ochotona_princeps | ENSOPRP00000014518 | 98.913 | 97.2826 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | delphinapterus_leucas | ENSDLEP00000005762 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | physeter_catodon | ENSPCTP00005025666 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 96.7391 | mus_caroli | MGP_CAROLIEiJ_P0087993 | 98.913 | 96.7391 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 96.1957 | mus_musculus | ENSMUSP00000034827 | 98.913 | 96.1957 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.3696 | 95.1087 | dipodomys_ordii | ENSDORP00000019218 | 98.3696 | 95.1087 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 98.3696 | mustela_putorius_furo | ENSMPUP00000020013 | 98.913 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.913 | aotus_nancymaae | ENSANAP00000002025 | 99.4565 | 98.913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 84.7826 | 82.6087 | aotus_nancymaae | ENSANAP00000038862 | 93.4132 | 91.018 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.0435 | 84.7826 | saimiri_boliviensis_boliviensis | ENSSBOP00000019616 | 94.186 | 90.6977 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.913 | saimiri_boliviensis_boliviensis | ENSSBOP00000001316 | 99.4565 | 98.913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 100 | 100 | chlorocebus_sabaeus | ENSCSAP00000019227 | 100 | 100 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.3696 | 96.7391 | mesocricetus_auratus | ENSMAUP00000008992 | 98.3696 | 96.7391 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 98.3696 | camelus_dromedarius | ENSCDRP00005005461 | 98.913 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.913 | carlito_syrichta | ENSTSYP00000033845 | 99.4565 | 98.913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 97.8261 | prolemur_simus | ENSPSMP00000037955 | 98.913 | 97.8261 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.3696 | 96.1957 | microtus_ochrogaster | ENSMOCP00000008627 | 98.3696 | 96.1957 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | sorex_araneus | ENSSARP00000008715 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 96.7391 | peromyscus_maniculatus_bairdii | ENSPEMP00000012764 | 98.913 | 96.7391 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | bos_mutus | ENSBMUP00000002381 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 96.1957 | mus_spicilegus | ENSMSIP00000010642 | 98.913 | 96.1957 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.3696 | 96.1957 | cricetulus_griseus_chok1gshd | ENSCGRP00001001550 | 98.3696 | 96.1957 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | pteropus_vampyrus | ENSPVAP00000009463 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 98.3696 | myotis_lucifugus | ENSMLUP00000012763 | 98.913 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 92.9348 | 85.3261 | vombatus_ursinus | ENSVURP00010015958 | 92.9348 | 85.3261 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | rhinolophus_ferrumequinum | ENSRFEP00010000296 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.913 | neovison_vison | ENSNVIP00000017361 | 99.4565 | 98.913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.3696 | 97.2826 | heterocephalus_glaber_female | ENSHGLP00000051464 | 98.3696 | 97.2826 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 100 | 99.4565 | rhinopithecus_bieti | ENSRBIP00000005577 | 100 | 99.4565 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 97.8261 | 96.1957 | canis_lupus_dingo | ENSCAFP00020004547 | 97.8261 | 96.1957 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 76.087 | 73.913 | canis_lupus_dingo | ENSCAFP00020008315 | 80.9249 | 78.6127 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.913 | sciurus_vulgaris | ENSSVLP00005025325 | 99.4565 | 98.913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.3696 | 97.2826 | procavia_capensis | ENSPCAP00000006423 | 98.3696 | 97.2826 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.3696 | 95.1087 | nannospalax_galili | ENSNGAP00000016539 | 98.3696 | 95.1087 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 94.5652 | 92.3913 | bos_grunniens | ENSBGRP00000037637 | 88.7755 | 86.7347 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | cervus_hanglu_yarkandensis | ENSCHYP00000020157 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 99.4565 | catagonus_wagneri | ENSCWAP00000011702 | 99.4565 | 99.4565 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 98.3696 | ictidomys_tridecemlineatus | ENSSTOP00000026497 | 98.913 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | otolemur_garnettii | ENSOGAP00000004389 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 85.8696 | 82.0652 | cebus_imitator | ENSCCAP00000011246 | 88.764 | 84.8315 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.913 | cebus_imitator | ENSCCAP00000000466 | 99.4565 | 98.913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.3696 | 97.8261 | urocitellus_parryii | ENSUPAP00010007228 | 98.3696 | 97.8261 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.3696 | moschus_moschiferus | ENSMMSP00000026652 | 99.4565 | 98.3696 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 100 | 99.4565 | rhinopithecus_roxellana | ENSRROP00000005984 | 100 | 99.4565 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 98.913 | 96.7391 | mus_pahari | MGP_PahariEiJ_P0023640 | 98.913 | 96.7391 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 99.4565 | 98.913 | propithecus_coquereli | ENSPCOP00000004194 | 99.4565 | 98.913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 66.8478 | 48.3696 | caenorhabditis_elegans | C48B6.2.1 | 67.2131 | 48.6339 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 76.087 | 55.4348 | drosophila_melanogaster | FBpp0086096 | 77.3481 | 56.3536 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 80.9783 | 57.0652 | ciona_intestinalis | ENSCINP00000008737 | 82.3204 | 58.0111 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 80.9783 | 61.9565 | eptatretus_burgeri | ENSEBUP00000003171 | 81.4208 | 62.2951 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 69.0217 | callorhinchus_milii | ENSCMIP00000008514 | 89.6175 | 69.3989 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 91.8478 | 72.8261 | latimeria_chalumnae | ENSLACP00000023320 | 92.3497 | 73.224 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.587 | 69.0217 | lepisosteus_oculatus | ENSLOCP00000012537 | 89.071 | 69.3989 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 80.4348 | 56.5217 | clupea_harengus | ENSCHAP00000014592 | 67.5799 | 47.4886 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.6739 | 67.3913 | danio_rerio | ENSDARP00000148639 | 90.1639 | 67.7596 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 68.4783 | carassius_auratus | ENSCARP00000094283 | 89.6175 | 68.8525 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 85.8696 | 65.2174 | astyanax_mexicanus | ENSAMXP00000019277 | 89.7727 | 68.1818 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 85.3261 | 65.7609 | ictalurus_punctatus | ENSIPUP00000019367 | 77.7228 | 59.901 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.587 | 67.3913 | esox_lucius | ENSELUP00000048250 | 89.071 | 67.7596 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.6739 | 66.8478 | electrophorus_electricus | ENSEEEP00000020993 | 90.1639 | 67.2131 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 86.9565 | 67.9348 | oncorhynchus_kisutch | ENSOKIP00005058907 | 87.4317 | 68.306 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 86.9565 | 67.9348 | oncorhynchus_mykiss | ENSOMYP00000007982 | 87.4317 | 68.306 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 86.413 | 68.4783 | salmo_salar | ENSSSAP00000090425 | 86.8852 | 68.8525 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 86.413 | 68.4783 | salmo_trutta | ENSSTUP00000079189 | 86.8852 | 68.8525 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.587 | 65.2174 | gadus_morhua | ENSGMOP00000019541 | 89.071 | 65.5738 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.0435 | 68.4783 | fundulus_heteroclitus | ENSFHEP00000015329 | 88.5246 | 68.8525 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 67.9348 | poecilia_reticulata | ENSPREP00000010107 | 89.6175 | 68.306 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 67.9348 | xiphophorus_maculatus | ENSXMAP00000025494 | 89.6175 | 68.306 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 69.0217 | oryzias_latipes | ENSORLP00000005632 | 89.6175 | 69.3989 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.0435 | 68.4783 | cyclopterus_lumpus | ENSCLMP00005016909 | 88.5246 | 68.8525 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 69.0217 | oreochromis_niloticus | ENSONIP00000043676 | 89.6175 | 69.3989 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.6739 | 69.5652 | haplochromis_burtoni | ENSHBUP00000016476 | 90.1639 | 69.9454 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.6739 | 69.5652 | astatotilapia_calliptera | ENSACLP00000014575 | 90.1639 | 69.9454 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.587 | 68.4783 | sparus_aurata | ENSSAUP00010043614 | 89.071 | 68.8525 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 67.3913 | lates_calcarifer | ENSLCAP00010051375 | 89.6175 | 67.7596 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.587 | 66.8478 | xenopus_tropicalis | ENSXETP00000100976 | 92.6136 | 69.8864 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 85.3261 | 73.3696 | chrysemys_picta_bellii | ENSCPBP00000030805 | 84.8649 | 72.973 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 75.5435 | 65.7609 | crocodylus_porosus | ENSCPRP00005000917 | 46.8013 | 40.7407 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 75 | laticauda_laticaudata | ENSLLTP00000020614 | 89.1304 | 75 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.587 | 74.4565 | notechis_scutatus | ENSNSUP00000026913 | 87.1658 | 73.262 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.587 | 73.913 | pseudonaja_textilis | ENSPTXP00000001770 | 88.587 | 73.913 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 80.9783 | 64.6739 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000022942 | 80.9783 | 64.6739 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 82.6087 | 66.8478 | gallus_gallus | ENSGALP00010015680 | 82.1622 | 66.4865 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 56.5217 | 41.8478 | serinus_canaria | ENSSCAP00000019531 | 69.3333 | 51.3333 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 83.6957 | 65.2174 | parus_major | ENSPMJP00000017231 | 83.2432 | 64.8649 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 90.2174 | 74.4565 | anolis_carolinensis | ENSACAP00000015973 | 54.6053 | 45.0658 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 79.3478 | 58.1522 | neolamprologus_brichardi | ENSNBRP00000005098 | 78.0749 | 57.2192 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.587 | 74.4565 | naja_naja | ENSNNAP00000007983 | 88.587 | 74.4565 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 73.913 | 60.8696 | podarcis_muralis | ENSPMRP00000002568 | 58.1197 | 47.8632 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 84.2391 | 65.2174 | taeniopygia_guttata | ENSTGUP00000013619 | 83.7838 | 64.8649 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 70.1087 | erpetoichthys_calabaricus | ENSECRP00000032379 | 89.6175 | 70.4918 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.0435 | 67.3913 | kryptolebias_marmoratus | ENSKMAP00000002555 | 88.5246 | 67.7596 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.0435 | 75 | salvator_merianae | ENSSMRP00000012225 | 86.631 | 73.7968 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 86.9565 | 65.2174 | oryzias_javanicus | ENSOJAP00000025023 | 81.2183 | 60.9137 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 86.413 | 65.2174 | amphilophus_citrinellus | ENSACIP00000028505 | 85.0267 | 64.1711 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 73.3696 | 53.2609 | dicentrarchus_labrax | ENSDLAP00005002700 | 54.4355 | 39.5161 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 66.3043 | amphiprion_percula | ENSAPEP00000007292 | 89.6175 | 66.6667 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 69.0217 | oryzias_sinensis | ENSOSIP00000008103 | 89.6175 | 69.3989 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 86.413 | 74.4565 | gopherus_evgoodei | ENSGEVP00005015151 | 85.9459 | 74.0541 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 85.3261 | 72.8261 | chelonoidis_abingdonii | ENSCABP00000016309 | 84.8649 | 72.4324 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.6739 | 69.0217 | seriola_dumerili | ENSSDUP00000014508 | 90.1639 | 69.3989 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 84.2391 | 62.5 | cottoperca_gobio | ENSCGOP00000022396 | 79.8969 | 59.2784 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 65.2174 | 53.8043 | anser_brachyrhynchus | ENSABRP00000002939 | 69.7674 | 57.5581 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.587 | 66.8478 | gasterosteus_aculeatus | ENSGACP00000021111 | 89.071 | 67.2131 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 66.8478 | cynoglossus_semilaevis | ENSCSEP00000026926 | 89.6175 | 67.2131 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 68.4783 | poecilia_formosa | ENSPFOP00000016606 | 89.6175 | 68.8525 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.6739 | 67.9348 | myripristis_murdjan | ENSMMDP00005032596 | 90.1639 | 68.306 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 90.2174 | 69.0217 | sander_lucioperca | ENSSLUP00000014972 | 90.7104 | 69.3989 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 82.0652 | 62.5 | strigops_habroptila | ENSSHBP00005012672 | 81.6216 | 62.1622 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 90.7609 | 67.3913 | amphiprion_ocellaris | ENSAOCP00000011464 | 91.2568 | 67.7596 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 86.413 | 66.8478 | hucho_hucho | ENSHHUP00000063085 | 86.8852 | 67.2131 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 57.0652 | 40.7609 | hucho_hucho | ENSHHUP00000035638 | 64.8148 | 46.2963 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.0435 | 65.7609 | betta_splendens | ENSBSLP00000047155 | 88.5246 | 66.1202 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 90.2174 | 69.0217 | pygocentrus_nattereri | ENSPNAP00000011464 | 90.7104 | 69.3989 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 87.5 | 64.6739 | anabas_testudineus | ENSATEP00000029041 | 76.6667 | 56.6667 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 80.4348 | 60.3261 | ciona_savignyi | ENSCSAVP00000001254 | 81.768 | 61.326 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 71.1957 | 51.087 | saccharomyces_cerevisiae | YHR148W | 71.5847 | 51.3661 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.6739 | 67.9348 | seriola_lalandi_dorsalis | ENSSLDP00000019901 | 90.1639 | 68.306 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.587 | 66.8478 | labrus_bergylta | ENSLBEP00000010584 | 89.071 | 67.2131 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.6739 | 69.5652 | pundamilia_nyererei | ENSPNYP00000000417 | 90.1639 | 69.9454 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 86.413 | 65.2174 | mastacembelus_armatus | ENSMAMP00000045687 | 76.0766 | 57.4163 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 90.7609 | 67.9348 | stegastes_partitus | ENSSPAP00000002726 | 91.2568 | 68.306 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 86.9565 | 67.9348 | oncorhynchus_tshawytscha | ENSOTSP00005002132 | 87.4317 | 68.306 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.6739 | 66.3043 | acanthochromis_polyacanthus | ENSAPOP00000012100 | 90.1639 | 66.6667 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 67.3913 | 52.7174 | aquila_chrysaetos_chrysaetos | ENSACCP00020011991 | 64.2487 | 50.2591 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 84.7826 | 65.2174 | nothobranchius_furzeri | ENSNFUP00015030661 | 81.6754 | 62.8272 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 84.7826 | 62.5 | cyprinodon_variegatus | ENSCVAP00000030744 | 84.3243 | 62.1622 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 87.5 | 65.2174 | denticeps_clupeoides | ENSDCDP00000008945 | 87.9781 | 65.5738 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 90.2174 | 69.0217 | larimichthys_crocea | ENSLCRP00005032266 | 90.7104 | 69.3989 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 86.413 | 65.2174 | takifugu_rubripes | ENSTRUP00000065466 | 72.2727 | 54.5455 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 64.1304 | 47.2826 | ficedula_albicollis | ENSFALP00000015426 | 63.4409 | 46.7742 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 86.413 | 63.587 | hippocampus_comes | ENSHCOP00000028141 | 85.4839 | 62.9032 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 70.1087 | 47.8261 | cyprinus_carpio_carpio | ENSCCRP00000006265 | 79.6296 | 54.321 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 67.9348 | cyprinus_carpio_carpio | ENSCCRP00000006249 | 89.6175 | 68.306 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 82.6087 | 66.3043 | coturnix_japonica | ENSCJPP00005002510 | 82.1622 | 65.9459 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 68.4783 | poecilia_latipinna | ENSPLAP00000026219 | 89.6175 | 68.8525 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 89.1304 | 67.9348 | sinocyclocheilus_grahami | ENSSGRP00000108675 | 89.6175 | 68.306 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 79.8913 | 58.1522 | maylandia_zebra | ENSMZEP00005007205 | 78.6096 | 57.2192 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 88.587 | 69.0217 | paramormyrops_kingsleyae | ENSPKIP00000023636 | 89.071 | 69.3989 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 90.2174 | 69.0217 | scleropages_formosus | ENSSFOP00015019402 | 90.7104 | 69.3989 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 77.7174 | 53.2609 | leptobrachium_leishanense | ENSLLEP00000038900 | 47.6667 | 32.6667 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 90.2174 | 68.4783 | scophthalmus_maximus | ENSSMAP00000012768 | 90.7104 | 68.8525 |
| ENSG00000177971 | homo_sapiens | ENSP00000385217 | 84.2391 | 64.1304 | oryzias_melastigma | ENSOMEP00000014632 | 84.2391 | 64.1304 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0003735 | enables structural constituent of ribosome | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005730 | located_in nucleolus | 2 | IDA,NAS | Homo_sapiens(9606) | Component |
| GO:0006364 | involved_in rRNA processing | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0015935 | part_of small ribosomal subunit | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0019843 | enables rRNA binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0030490 | involved_in maturation of SSU-rRNA | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0030515 | enables snoRNA binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0030684 | part_of preribosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0032040 | part_of small-subunit processome | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0034457 | part_of Mpp10 complex | 3 | IBA,IDA,IPI | Homo_sapiens(9606) | Component |
| GO:0042274 | involved_in ribosomal small subunit biogenesis | 2 | IBA,IDA | Homo_sapiens(9606) | Process |