RBP Type
Non-canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
Kinase
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000174672 Gene ID
9024 Accession
11405
Symbol
BRSK2
Alias
SAD1;SADA;SAD-A;STK29;PEN11B;C11orf7
Full Name
BR serine/threonine kinase 2
Status
Confidence
Length
72791 bases
Strand
Plus strand
Position
11 : 1389899 - 1462689
RNA binding domain
N.A.
Summary
Enables several functions, including ATP binding activity; ATPase binding activity; and protein kinase activity. Involved in several processes, including cellular protein metabolic process; intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress; and regulation of insulin secretion involved in cellular response to glucose stimulus. Located in centrosome and endoplasmic reticulum. [provided by Alliance of Genome Resources, Apr 2022]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
BRSK2-201
ENST00000308219
4089
668aa
ENSP00000310697
BRSK2-209
ENST00000529433
3211
674aa
ENSP00000433684
BRSK2-208
ENST00000528841
4528
736aa
ENSP00000432000
BRSK2-212
ENST00000531197
3163
674aa
ENSP00000431152
BRSK2-204
ENST00000526678
2091
696aa
ENSP00000433370
BRSK2-206
ENST00000528596
581
86aa
ENSP00000434075
BRSK2-203
ENST00000524702
580
130aa
ENSP00000432672
BRSK2-207
ENST00000528710
3528
614aa
ENSP00000433235
BRSK2-202
ENST00000382179
3576
766aa
ENSP00000371614
BRSK2-210
ENST00000529951
1934
170aa
ENSP00000432252
BRSK2-211
ENST00000531078
600
No protein
-
BRSK2-214
ENST00000533606
2319
200aa
ENSP00000436474
BRSK2-205
ENST00000526768
1956
No protein
-
BRSK2-213
ENST00000531932
709
No protein
-
BRSK2-215
ENST00000544817
1842
170aa
ENSP00000445168
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
30360 Potassium 4.4380000E-005 - 79815 Heroin Dependence 4E-6 27936112
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
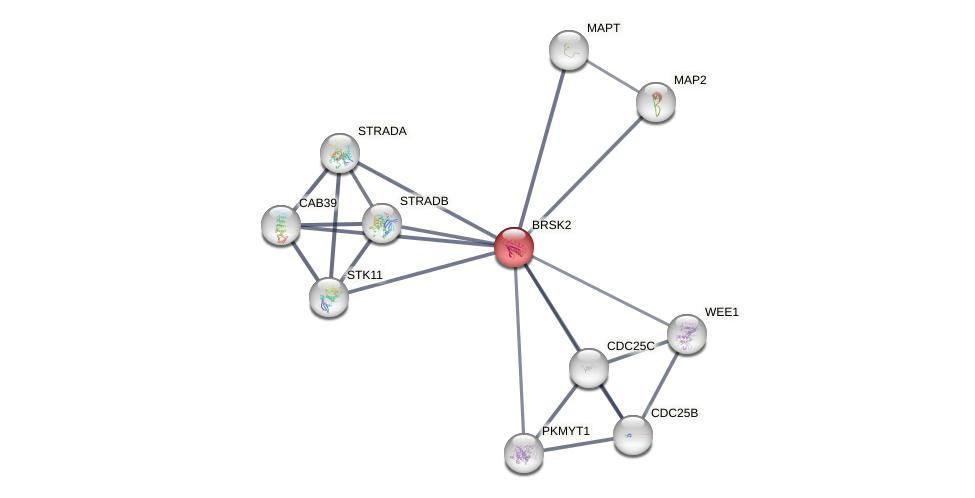
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000086 involved_in G2/M transition of mitotic cell cycle 1 IMP Homo_sapiens(9606) Process GO:0000287 enables magnesium ion binding 1 IDA Homo_sapiens(9606) Function GO:0004674 enables protein serine/threonine kinase activity 3 IBA,IDA,ISS Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005524 enables ATP binding 1 IDA Homo_sapiens(9606) Function GO:0005737 located_in cytoplasm 1 IDA Homo_sapiens(9606) Component GO:0005783 located_in endoplasmic reticulum 1 IDA Homo_sapiens(9606) Component GO:0005813 is_active_in centrosome 2 IBA,IDA Homo_sapiens(9606) Component GO:0006468 involved_in protein phosphorylation 2 IBA,IDA Homo_sapiens(9606) Process GO:0006887 involved_in exocytosis 1 IEA Homo_sapiens(9606) Process GO:0007409 involved_in axonogenesis 2 IBA,ISS Homo_sapiens(9606) Process GO:0010975 involved_in regulation of neuron projection development 1 ISS Homo_sapiens(9606) Process GO:0018105 involved_in peptidyl-serine phosphorylation 1 IEA Homo_sapiens(9606) Process GO:0019901 enables protein kinase binding 1 IEA Homo_sapiens(9606) Function GO:0030010 involved_in establishment of cell polarity 2 IBA,ISS Homo_sapiens(9606) Process GO:0030036 involved_in actin cytoskeleton organization 1 IMP Homo_sapiens(9606) Process GO:0030182 involved_in neuron differentiation 1 ISS Homo_sapiens(9606) Process GO:0035556 involved_in intracellular signal transduction 1 IBA Homo_sapiens(9606) Process GO:0036503 involved_in ERAD pathway 1 IMP Homo_sapiens(9606) Process GO:0043462 involved_in regulation of ATP-dependent activity 1 NAS Homo_sapiens(9606) Process GO:0048156 enables tau protein binding 1 NAS Homo_sapiens(9606) Function GO:0048471 located_in perinuclear region of cytoplasm 1 IEA Homo_sapiens(9606) Component GO:0050321 enables tau-protein kinase activity 4 IBA,IDA,ISS,NAS Homo_sapiens(9606) Function GO:0050770 involved_in regulation of axonogenesis 1 ISS Homo_sapiens(9606) Process GO:0051117 enables ATPase binding 1 IDA Homo_sapiens(9606) Function GO:0051301 involved_in cell division 1 IEA Homo_sapiens(9606) Process GO:0060590 enables ATPase regulator activity 1 NAS Homo_sapiens(9606) Function GO:0061178 involved_in regulation of insulin secretion involved in cellular response to glucose stimulus 2 IMP,ISS Homo_sapiens(9606) Process GO:0070059 involved_in intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress 1 IMP Homo_sapiens(9606) Process GO:0090176 involved_in microtubule cytoskeleton organization involved in establishment of planar polarity 1 ISS Homo_sapiens(9606) Process GO:0106310 enables protein serine kinase activity 1 IEA Homo_sapiens(9606) Function GO:0150034 located_in distal axon 1 ISS Homo_sapiens(9606) Component GO:1904152 involved_in regulation of retrograde protein transport, ER to cytosol 2 IC,NAS Homo_sapiens(9606) Process GO:2000807 involved_in regulation of synaptic vesicle clustering 1 TAS Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.