RBP Type
Canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
E3 ligase
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000174013 Gene ID
200933 Accession
29148
Symbol
FBXO45
Alias
Fbx45
Full Name
F-box protein 45
Status
Confidence
Length
20449 bases
Strand
Plus strand
Position
3 : 196568611 - 196589059
RNA binding domain
SPRY
Summary
Members of the F-box protein family, such as FBXO45, are characterized by an approximately 40-amino acid F-box motif. SCF complexes, formed by SKP1 (MIM 601434), cullin (see CUL1; MIM 603134), and F-box proteins, act as protein-ubiquitin ligases. F-box proteins interact with SKP1 through the F box, and they interact with ubiquitination targets through other protein interaction domains (summary by Jin et al., 2004 [PubMed 15520277]).[supplied by OMIM, Jan 2011]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000310332
SPRY
PF00622 7.8e-18
1
ENSP00000389868
SPRY
PF00622 6.7e-13
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
FBXO45-202
ENST00000440469
3689
107aa
ENSP00000389868
FBXO45-201
ENST00000311630
5927
286aa
ENSP00000310332
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
33033 Ankle Brachial Index 5.9973403E-005 -
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
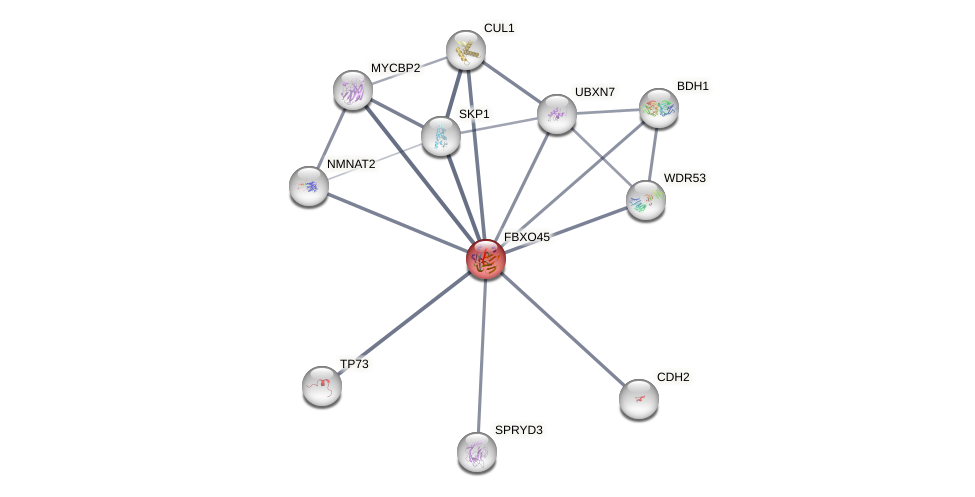
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0001764 involved_in neuron migration 1 IEA Homo_sapiens(9606) Process GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005576 located_in extracellular region 1 IEA Homo_sapiens(9606) Component GO:0005634 located_in nucleus 1 IEA Homo_sapiens(9606) Component GO:0005737 is_active_in cytoplasm 1 IC Homo_sapiens(9606) Component GO:0006511 involved_in ubiquitin-dependent protein catabolic process 1 IDA Homo_sapiens(9606) Process GO:0006974 involved_in DNA damage response 1 IDA Homo_sapiens(9606) Process GO:0014069 located_in postsynaptic density 1 IEA Homo_sapiens(9606) Component GO:0016567 involved_in protein ubiquitination 2 IDA,ISS Homo_sapiens(9606) Process GO:0019005 part_of SCF ubiquitin ligase complex 1 IBA Homo_sapiens(9606) Component GO:0021799 involved_in cerebral cortex radially oriented cell migration 1 IEA Homo_sapiens(9606) Process GO:0021800 involved_in cerebral cortex tangential migration 1 IEA Homo_sapiens(9606) Process GO:0021957 involved_in corticospinal tract morphogenesis 1 IEA Homo_sapiens(9606) Process GO:0021960 involved_in anterior commissure morphogenesis 1 IEA Homo_sapiens(9606) Process GO:0042734 located_in presynaptic membrane 1 IEA Homo_sapiens(9606) Component GO:0042995 located_in cell projection 1 IEA Homo_sapiens(9606) Component GO:0043161 involved_in proteasome-mediated ubiquitin-dependent protein catabolic process 2 IBA,ISS Homo_sapiens(9606) Process GO:0045202 is_active_in synapse 1 IBA Homo_sapiens(9606) Component GO:0045211 located_in postsynaptic membrane 1 IEA Homo_sapiens(9606) Component GO:0060386 involved_in synapse assembly involved in innervation 1 IBA Homo_sapiens(9606) Process GO:0070936 involved_in protein K48-linked ubiquitination 1 IDA Homo_sapiens(9606) Process GO:0098978 located_in glutamatergic synapse 1 IEA Homo_sapiens(9606) Component GO:0099523 located_in presynaptic cytosol 1 IEA Homo_sapiens(9606) Component GO:0099524 located_in postsynaptic cytosol 1 IEA Homo_sapiens(9606) Component GO:1990756 enables ubiquitin ligase-substrate adaptor activity 1 IDA Homo_sapiens(9606) Function GO:2000300 involved_in regulation of synaptic vesicle exocytosis 1 IEA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.