Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000172977 | Gene ID | 10524 | Accession | 5275 |
| Symbol | KAT5 | Alias | TIP;ESA1;PLIP;TIP60;cPLA2;HTATIP;ZC2HC5;HTATIP1;NEDFASB | Full Name | lysine acetyltransferase 5 |
| Status | Confidence | Length | 7609 bases | Strand | Plus strand |
| Position | 11 : 65711996 - 65719604 | RNA binding domain | Tudor-knot | ||
| Summary | The protein encoded by this gene belongs to the MYST family of histone acetyl transferases (HATs) and was originally isolated as an HIV-1 TAT-interactive protein. HATs play important roles in regulating chromatin remodeling, transcription and other nuclear processes by acetylating histone and nonhistone proteins. This protein is a histone acetylase that has a role in DNA repair and apoptosis and is thought to play an important role in signal transduction. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000436247 | Tudor-knot | PF11717 | 6.3e-19 | 1 |
| ENSP00000344955 | Tudor-knot | PF11717 | 3.6e-18 | 1 |
| ENSP00000434765 | Tudor-knot | PF11717 | 4e-18 | 1 |
| ENSP00000366245 | Tudor-knot | PF11717 | 4e-18 | 1 |
| ENSP00000340330 | Tudor-knot | PF11717 | 4.5e-18 | 1 |
| ENSP00000436000 | Tudor-knot | PF11717 | 3.6e-15 | 1 |
| ENSP00000436012 | Tudor-knot | PF11717 | 6.1e-15 | 1 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| KAT5-203 | ENST00000377046 | 2237 | 513aa | ENSP00000366245 |
| KAT5-202 | ENST00000352980 | 2059 | 461aa | ENSP00000344955 |
| KAT5-211 | ENST00000532042 | 763 | No protein | - |
| KAT5-208 | ENST00000530446 | 1697 | 494aa | ENSP00000434765 |
| KAT5-214 | ENST00000534104 | 576 | 16aa | ENSP00000435939 |
| KAT5-201 | ENST00000341318 | 2080 | 546aa | ENSP00000340330 |
| KAT5-209 | ENST00000530605 | 499 | 167aa | ENSP00000436247 |
| KAT5-215 | ENST00000534293 | 559 | No protein | - |
| KAT5-206 | ENST00000527544 | 745 | No protein | - |
| KAT5-207 | ENST00000528198 | 609 | 189aa | ENSP00000436000 |
| KAT5-210 | ENST00000531880 | 845 | 269aa | ENSP00000436012 |
| KAT5-216 | ENST00000534650 | 1875 | 302aa | ENSP00000431819 |
| KAT5-204 | ENST00000525204 | 574 | No protein | - |
| KAT5-217 | ENST00000534681 | 583 | 45aa | ENSP00000434993 |
| KAT5-212 | ENST00000533441 | 735 | No protein | - |
| KAT5-205 | ENST00000525600 | 512 | No protein | - |
| KAT5-213 | ENST00000533596 | 709 | 95aa | ENSP00000435766 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
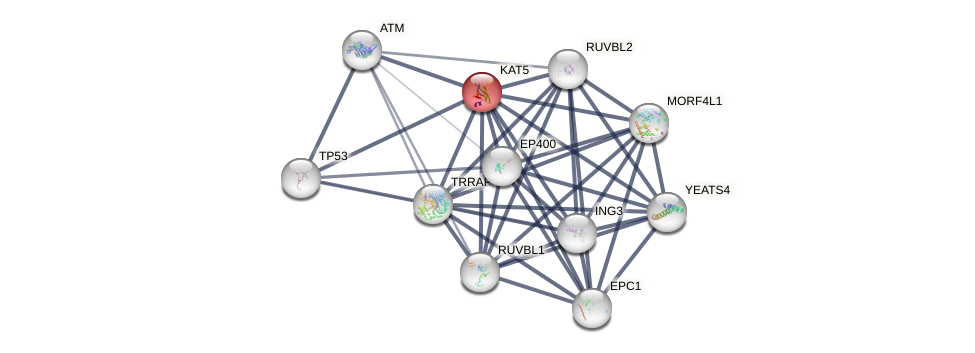
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000172977 | homo_sapiens | ENSP00000219797 | 48.2533 | 37.3362 | homo_sapiens | ENSP00000340330 | 40.4762 | 31.3187 |
| ENSG00000172977 | homo_sapiens | ENSP00000347716 | 24.031 | 14.9871 | homo_sapiens | ENSP00000340330 | 17.033 | 10.6227 |
| ENSG00000172977 | homo_sapiens | ENSP00000450518 | 25.3968 | 14.8148 | homo_sapiens | ENSP00000340330 | 17.5824 | 10.2564 |
| ENSG00000172977 | homo_sapiens | ENSP00000436901 | 22.5064 | 13.2992 | homo_sapiens | ENSP00000340330 | 16.1172 | 9.52381 |
| ENSG00000172977 | homo_sapiens | ENSP00000287239 | 14.1823 | 10.0338 | homo_sapiens | ENSP00000340330 | 53.8462 | 38.0952 |
| ENSG00000172977 | homo_sapiens | ENSP00000259021 | 39.6072 | 29.2962 | homo_sapiens | ENSP00000340330 | 44.3223 | 32.7839 |
| ENSG00000172977 | homo_sapiens | ENSP00000341805 | 19.4779 | 10.0402 | homo_sapiens | ENSP00000340330 | 17.7656 | 9.15751 |
| ENSG00000172977 | homo_sapiens | ENSP00000265713 | 14.6208 | 10.3792 | homo_sapiens | ENSP00000340330 | 53.663 | 38.0952 |
| ENSG00000172977 | homo_sapiens | ENSP00000311513 | 10.4094 | 5.41291 | homo_sapiens | ENSP00000340330 | 27.4725 | 14.2857 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 97.619 | 97.619 | nomascus_leucogenys | ENSNLEP00000037048 | 99.6262 | 99.6262 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.956 | pan_paniscus | ENSPPAP00000038731 | 100 | 100 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 100 | 100 | pongo_abelii | ENSPPYP00000035578 | 100 | 100 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.956 | pan_troglodytes | ENSPTRP00000006709 | 100 | 100 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.956 | gorilla_gorilla | ENSGGOP00000023278 | 100 | 100 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 97.619 | 96.1538 | ornithorhynchus_anatinus | ENSOANP00000036523 | 97.2628 | 95.8029 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.7729 | 92.8571 | sarcophilus_harrisii | ENSSHAP00000002166 | 98.0843 | 97.1264 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 88.8278 | 88.0952 | notamacropus_eugenii | ENSMEUP00000005661 | 99.7942 | 98.9712 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 43.0403 | 42.4908 | erinaceus_europaeus | ENSEEUP00000011699 | 48.8565 | 48.2328 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.956 | callithrix_jacchus | ENSCJAP00000003663 | 100 | 100 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 88.4615 | 85.8974 | cercocebus_atys | ENSCATP00000030502 | 89.9441 | 87.3371 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.7729 | 93.7729 | macaca_fascicularis | ENSMFAP00000008718 | 99.6109 | 99.6109 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.7729 | 93.7729 | macaca_mulatta | ENSMMUP00000026085 | 99.6109 | 99.6109 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.6337 | macaca_nemestrina | ENSMNEP00000023759 | 99.6337 | 99.6337 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.8168 | papio_anubis | ENSPANP00000005348 | 99.8168 | 99.8168 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.8168 | mandrillus_leucophaeus | ENSMLEP00000021669 | 99.8168 | 99.8168 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | vulpes_vulpes | ENSVVUP00000037884 | 98.018 | 97.8378 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.7729 | mus_spretus | MGP_SPRETEiJ_P0049606 | 100 | 99.8051 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | equus_asinus | ENSEASP00005053850 | 99.6337 | 99.4505 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | capra_hircus | ENSCHIP00000014439 | 99.6337 | 99.4505 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.2674 | 99.0843 | loxodonta_africana | ENSLAFP00000013105 | 99.4495 | 99.2661 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | ailuropoda_melanoleuca | ENSAMEP00000018343 | 99.6337 | 99.4505 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.7729 | 93.5897 | bos_indicus_hybrid | ENSBIXP00005012074 | 99.8051 | 99.6101 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 88.2784 | 86.8132 | oryctolagus_cuniculus | ENSOCUP00000038189 | 77.7419 | 76.4516 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | equus_caballus | ENSECAP00000027492 | 98.3725 | 98.1917 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | canis_lupus_familiaris | ENSCAFP00845026127 | 98.018 | 97.8378 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.7729 | panthera_pardus | ENSPPRP00000006832 | 100 | 99.8051 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 91.2088 | 89.5604 | marmota_marmota_marmota | ENSMMMP00000023994 | 90.8759 | 89.2336 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.6337 | balaenoptera_musculus | ENSBMSP00010013117 | 99.8168 | 99.6337 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.5897 | cavia_porcellus | ENSCPOP00000022456 | 94.8244 | 94.4547 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | ursus_americanus | ENSUAMP00000026884 | 99.6337 | 99.4505 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 94.8718 | 93.4066 | monodelphis_domestica | ENSMODP00000011971 | 83.9546 | 82.658 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | bison_bison_bison | ENSBBBP00000027734 | 99.6337 | 99.4505 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.7729 | monodon_monoceros | ENSMMNP00015030020 | 100 | 99.8051 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 79.4872 | 74.5421 | sus_scrofa | ENSSSCP00000013803 | 78.7659 | 73.8657 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.6337 | microcebus_murinus | ENSMICP00000022678 | 99.8168 | 99.6337 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.5897 | 93.4066 | octodon_degus | ENSODEP00000024387 | 99.6101 | 99.4152 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.7729 | jaculus_jaculus | ENSJJAP00000024089 | 100 | 99.8051 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | ovis_aries_rambouillet | ENSOARP00020026655 | 99.6337 | 99.4505 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.6337 | delphinapterus_leucas | ENSDLEP00000004290 | 99.8168 | 99.6337 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.7729 | physeter_catodon | ENSPCTP00005026794 | 93.956 | 93.7729 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.7729 | mus_caroli | MGP_CAROLIEiJ_P0047644 | 100 | 99.8051 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.7729 | mus_musculus | ENSMUSP00000109271 | 100 | 99.8051 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.5897 | dipodomys_ordii | ENSDORP00000011811 | 100 | 99.6101 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.7729 | mustela_putorius_furo | ENSMPUP00000011528 | 100 | 99.8051 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.8168 | aotus_nancymaae | ENSANAP00000035852 | 99.8168 | 99.8168 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.956 | saimiri_boliviensis_boliviensis | ENSSBOP00000007645 | 100 | 100 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.956 | chlorocebus_sabaeus | ENSCSAP00000004254 | 98.2759 | 98.2759 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.7729 | 93.4066 | mesocricetus_auratus | ENSMAUP00000017078 | 99.4175 | 99.0291 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.6337 | phocoena_sinus | ENSPSNP00000014764 | 99.8168 | 99.6337 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | camelus_dromedarius | ENSCDRP00005006425 | 99.6337 | 99.4505 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.6337 | carlito_syrichta | ENSTSYP00000025835 | 99.8168 | 99.6337 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | panthera_tigris_altaica | ENSPTIP00000012210 | 99.6337 | 99.4505 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.6337 | rattus_norvegicus | ENSRNOP00000095340 | 99.8168 | 99.6337 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.6337 | prolemur_simus | ENSPSMP00000033422 | 99.8168 | 99.6337 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.4066 | microtus_ochrogaster | ENSMOCP00000009062 | 99.8055 | 99.2218 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 89.9267 | 89.1941 | sorex_araneus | ENSSARP00000010510 | 90.0917 | 89.3578 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.7729 | peromyscus_maniculatus_bairdii | ENSPEMP00000015697 | 100 | 99.8051 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.7729 | 93.5897 | bos_mutus | ENSBMUP00000001351 | 99.8051 | 99.6101 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.6337 | mus_spicilegus | ENSMSIP00000001421 | 99.8168 | 99.6337 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.7729 | 93.2234 | cricetulus_griseus_chok1gshd | ENSCGRP00001015476 | 99.0329 | 98.4526 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.4505 | 99.0843 | pteropus_vampyrus | ENSPVAP00000015987 | 99.4505 | 99.0843 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.2234 | 93.0403 | myotis_lucifugus | ENSMLUP00000007984 | 99.2203 | 99.0253 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.7729 | 92.8571 | vombatus_ursinus | ENSVURP00010001917 | 99.8051 | 98.8304 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | rhinolophus_ferrumequinum | ENSRFEP00010005282 | 99.6337 | 99.4505 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | neovison_vison | ENSNVIP00000019941 | 99.6337 | 99.4505 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | bos_taurus | ENSBTAP00000070633 | 99.6337 | 99.4505 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 95.9707 | 95.2381 | heterocephalus_glaber_female | ENSHGLP00000013387 | 84.9271 | 84.2788 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.956 | rhinopithecus_bieti | ENSRBIP00000028673 | 100 | 100 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | canis_lupus_dingo | ENSCAFP00020018985 | 98.018 | 97.8378 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.7729 | sciurus_vulgaris | ENSSVLP00005012197 | 100 | 99.8051 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 87.5458 | 86.2637 | phascolarctos_cinereus | ENSPCIP00000052872 | 90.3592 | 89.0359 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.4066 | nannospalax_galili | ENSNGAP00000018581 | 95.8878 | 95.3271 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.7729 | chinchilla_lanigera | ENSCLAP00000023011 | 100 | 99.8051 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | cervus_hanglu_yarkandensis | ENSCHYP00000007475 | 99.6337 | 99.4505 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.6337 | catagonus_wagneri | ENSCWAP00000013810 | 99.8168 | 99.6337 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.6337 | ictidomys_tridecemlineatus | ENSSTOP00000028998 | 99.8168 | 99.6337 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.7729 | 93.5897 | otolemur_garnettii | ENSOGAP00000012652 | 99.8051 | 99.6101 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.8168 | cebus_imitator | ENSCCAP00000031184 | 99.8168 | 99.8168 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.6337 | urocitellus_parryii | ENSUPAP00010011630 | 98.1982 | 98.018 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.6337 | 99.4505 | moschus_moschiferus | ENSMMSP00000023068 | 99.6337 | 99.4505 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.8168 | rhinopithecus_roxellana | ENSRROP00000039022 | 99.8168 | 99.8168 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 93.7729 | mus_pahari | MGP_PahariEiJ_P0020655 | 100 | 99.8051 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 99.8168 | 99.6337 | propithecus_coquereli | ENSPCOP00000032196 | 99.8168 | 99.6337 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 11.9048 | 6.77656 | caenorhabditis_elegans | C28H8.9a.1 | 17.4731 | 9.94624 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 10.8059 | 7.87546 | drosophila_melanogaster | FBpp0311947 | 13.785 | 10.0467 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 13.0037 | 7.87546 | drosophila_melanogaster | FBpp0085418 | 14.2857 | 8.65191 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 38.0952 | 31.685 | ciona_intestinalis | ENSCINP00000031062 | 75.9124 | 63.1387 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 26.3736 | 24.1758 | callorhinchus_milii | ENSCMIP00000009052 | 77.4194 | 70.9677 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 79.8535 | 71.978 | latimeria_chalumnae | ENSLACP00000011674 | 84.9902 | 76.6082 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 56.4103 | 46.337 | gadus_morhua | ENSGMOP00000044570 | 40.052 | 32.8999 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.956 | 91.9414 | chrysemys_picta_bellii | ENSCPBP00000033470 | 92.9348 | 90.942 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 91.9414 | 89.5604 | sphenodon_punctatus | ENSSPUP00000001194 | 97.6654 | 95.1362 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 90.6593 | 88.2784 | laticauda_laticaudata | ENSLLTP00000009580 | 96.4912 | 93.9571 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 83.1502 | 80.5861 | pseudonaja_textilis | ENSPTXP00000014962 | 93.8017 | 90.9091 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 34.4322 | 28.5714 | gallus_gallus | ENSGALP00010001520 | 54.023 | 44.8276 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 61.7216 | 55.8608 | neolamprologus_brichardi | ENSNBRP00000027396 | 67.4 | 61 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 91.5751 | 88.8278 | podarcis_muralis | ENSPMRP00000023314 | 95.6023 | 92.7342 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 92.4908 | 90.4762 | pelodiscus_sinensis | ENSPSIP00000003037 | 98.0583 | 95.9223 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 92.3077 | 90.293 | gopherus_evgoodei | ENSGEVP00005022169 | 96.3671 | 94.2639 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 93.7729 | 91.5751 | chelonoidis_abingdonii | ENSCABP00000024090 | 94.4649 | 92.2509 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 92.1245 | 90.1099 | terrapene_carolina_triunguis | ENSTMTP00000026212 | 96.1759 | 94.0727 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 54.0293 | 38.8278 | saccharomyces_cerevisiae | YOR244W | 66.2921 | 47.6404 |
| ENSG00000172977 | homo_sapiens | ENSP00000340330 | 35.348 | 32.4176 | sinocyclocheilus_grahami | ENSSGRP00000049142 | 78.7755 | 72.2449 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000122 | involved_in negative regulation of transcription by RNA polymerase II | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0000132 | involved_in establishment of mitotic spindle orientation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0000724 | involved_in double-strand break repair via homologous recombination | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0000776 | located_in kinetochore | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0000785 | part_of chromatin | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0000786 | part_of nucleosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0000812 | part_of Swr1 complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0003682 | enables chromatin binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003712 | enables transcription coregulator activity | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0003713 | enables transcription coactivator activity | 2 | IDA,TAS | Homo_sapiens(9606) | Function |
| GO:0004402 | enables histone acetyltransferase activity | 2 | IDA,IMP | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 2 | IDA,IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 2 | IDA,NAS | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005667 | part_of transcription regulator complex | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005730 | located_in nucleolus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0006289 | involved_in nucleotide-excision repair | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006302 | involved_in double-strand break repair | 2 | IDA,TAS | Homo_sapiens(9606) | Process |
| GO:0006351 | involved_in DNA-templated transcription | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006915 | involved_in apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006974 | involved_in DNA damage response | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006978 | involved_in DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0007286 | involved_in spermatid development | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0010212 | involved_in response to ionizing radiation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0010485 | enables histone H4 acetyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0010508 | involved_in positive regulation of autophagy | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0010867 | involved_in positive regulation of triglyceride biosynthetic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0016407 | enables acetyltransferase activity | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0016573 | involved_in histone acetylation | 2 | IDA,NAS | Homo_sapiens(9606) | Process |
| GO:0018393 | involved_in internal peptidyl-lysine acetylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0018394 | involved_in peptidyl-lysine acetylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0021915 | involved_in neural tube development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0022008 | involved_in neurogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0032703 | involved_in negative regulation of interleukin-2 production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032777 | part_of Piccolo NuA4 histone acetyltransferase complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0035092 | involved_in sperm DNA condensation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0035267 | part_of NuA4 histone acetyltransferase complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0035861 | is_active_in site of double-strand break | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0042149 | involved_in cellular response to glucose starvation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0042753 | involved_in positive regulation of circadian rhythm | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0042981 | involved_in regulation of apoptotic process | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0043161 | involved_in proteasome-mediated ubiquitin-dependent protein catabolic process | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0043231 | located_in intracellular membrane-bounded organelle | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0043967 | involved_in histone H4 acetylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0043968 | involved_in histone H2A acetylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0043998 | enables histone H2A acetyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0043999 | enables histone H2AK5 acetyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0045087 | involved_in innate immune response | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045591 | involved_in positive regulation of regulatory T cell differentiation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045663 | involved_in positive regulation of myoblast differentiation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045892 | involved_in negative regulation of DNA-templated transcription | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0045893 | involved_in positive regulation of DNA-templated transcription | 2 | IDA,NAS | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 3 | IBA,IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0046872 | enables metal ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0046972 | enables histone H4K16 acetyltransferase activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0048471 | located_in perinuclear region of cytoplasm | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0051726 | involved_in regulation of cell cycle | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0061733 | enables peptide-lysine-N-acetyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0062033 | involved_in positive regulation of mitotic sister chromatid segregation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071333 | involved_in cellular response to glucose stimulus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071392 | involved_in cellular response to estradiol stimulus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0072487 | part_of MSL complex | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0090398 | involved_in cellular senescence | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0097431 | located_in mitotic spindle pole | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0106226 | enables peptide 2-hydroxyisobutyryltransferase activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0140064 | enables peptide crotonyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0140065 | enables peptide butyryltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0140297 | enables DNA-binding transcription factor binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0140861 | involved_in DNA repair-dependent chromatin remodeling | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1901985 | involved_in positive regulation of protein acetylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1902036 | involved_in regulation of hematopoietic stem cell differentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:1902425 | involved_in positive regulation of attachment of mitotic spindle microtubules to kinetochore | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1905168 | involved_in positive regulation of double-strand break repair via homologous recombination | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1905691 | acts_upstream_of_positive_effect lipid droplet disassembly | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:2000042 | involved_in negative regulation of double-strand break repair via homologous recombination | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:2000779 | involved_in regulation of double-strand break repair | 1 | NAS | Homo_sapiens(9606) | Process |