Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000164362 | Gene ID | 7015 | Accession | 11730 |
| Symbol | TERT | Alias | TP2;TRT;CMM9;EST2;TCS1;hTRT;DKCA2;DKCB4;hEST2;PFBMFT1 | Full Name | telomerase reverse transcriptase |
| Status | Confidence | Length | 41902 bases | Strand | Minus strand |
| Position | 5 : 1253167 - 1295068 | RNA binding domain | Telomerase_RBD , RVT_1 | ||
| Summary | Telomerase is a ribonucleoprotein polymerase that maintains telomere ends by addition of the telomere repeat TTAGGG. The enzyme consists of a protein component with reverse transcriptase activity, encoded by this gene, and an RNA component which serves as a template for the telomere repeat. Telomerase expression plays a role in cellular senescence, as it is normally repressed in postnatal somatic cells resulting in progressive shortening of telomeres. Deregulation of telomerase expression in somatic cells may be involved in oncogenesis. Studies in mouse suggest that telomerase also participates in chromosomal repair, since de novo synthesis of telomere repeats may occur at double-stranded breaks. Alternatively spliced variants encoding different isoforms of telomerase reverse transcriptase have been identified; the full-length sequence of some variants has not been determined. Alternative splicing at this locus is thought to be one mechanism of regulation of telomerase activity. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 31903880 | Telomere-related Markers for Cancer. | Xiaotian Yuan | 2020-01-01 | Current topics in medicinal chemistry |
| 11809815 | Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. | J Tara Moriarty | 2002-02-01 | Molecular and cellular biology |
| 37531400 | Dynamics of TERT regulation via alternative splicing in stem cells and cancer cells. | J Jeongjin Kim | 2023-01-01 | PloS one |
| 33553693 | Shelterin complex gene: Prognosis and therapeutic vulnerability in cancer. | Kumar Vikas Bhari | 2021-07-01 | Biochemistry and biophysics reports |
| 32242127 | Regulation of human telomerase in homeostasis and disease. | M Caitlin Roake | 2020-07-01 | Nature reviews. Molecular cell biology |
| 34474152 | Post-transcriptional diversity in riboproteins and RNAs in aging and cancer. | Jurandir Cruz | 2021-11-01 | Seminars in cancer biology |
| 22244753 | Telomerase and telomere-associated proteins: structural insights into mechanism and evolution. | A Karen Lewis | 2012-01-11 | Structure (London, England : 1993) |
| 35477117 | Functional interaction between compound heterozygous TERT mutations causes severe telomere biology disorder. | Aram Niaz | 2022-06-28 | Blood advances |
| 37009488 | Proteomic analysis defines the interactome of telomerase in the protozoan parasite, Trypanosoma brucei. | A Justin Davis | 2023-01-01 | Frontiers in cell and developmental biology |
| 28441946 | The protein subunit of telomerase displays patterns of dynamic evolution and conservation across different metazoan taxa. | G Alvina Lai | 2017-04-26 | BMC evolutionary biology |
| 23447707 | A translocation-defective telomerase with low levels of activity and processivity stabilizes short telomeres and confers immortalization. | Yasmin D'Souza | 2013-05-01 | Molecular biology of the cell |
| 28264499 | Current Insights to Regulation and Role of Telomerase in Human Diseases. | Burak Mert Ozturk | 2017-02-28 | Antioxidants (Basel, Switzerland) |
| 33046533 | A 4-Base-Pair Core-Enclosing Helix in Telomerase RNA Is Essential for Activity and for Binding to the Telomerase Reverse Transcriptase Catalytic Protein Subunit. | A Melissa Mefford | 2020-11-20 | Molecular and cellular biology |
| 34083519 | Nuclear compartmentalization of TERT mRNA and TUG1 lncRNA is driven by intron retention. | Gabrijela Dumbović | 2021-06-03 | Nature communications |
| 25684364 | A platform for rapid exploration of aging and diseases in a naturally short-lived vertebrate. | Itamar Harel | 2015-02-26 | Cell |
| 24957604 | Identification of RNA binding motifs in the R2 retrotransposon-encoded reverse transcriptase. | K Varuni Jamburuthugoda | 2014-07-01 | Nucleic acids research |
| 27359343 | Evolutionary perspectives of telomerase RNA structure and function. | D Joshua Podlevsky | 2016-08-02 | RNA biology |
| 23901009 | Many disease-associated variants of hTERT retain high telomerase enzymatic activity. | J Arthur Zaug | 2013-10-01 | Nucleic acids research |
| 23299958 | Finding the end: recruitment of telomerase to telomeres. | Jayakrishnan Nandakumar | 2013-02-01 | Nature reviews. Molecular cell biology |
| 31091669 | Insights into Telomerase/hTERT Alternative Splicing Regulation Using Bioinformatics and Network Analysis in Cancer. | T Andrew Ludlow | 2019-05-14 | Cancers |
| 31892146 | Oxoisoaporphines and Aporphines: Versatile Molecules with Anticancer Effects. | Esteban Rodríguez-Arce | 2019-12-27 | Molecules (Basel, Switzerland) |
| 22093366 | It all comes together at the ends: telomerase structure, function, and biogenesis. | D Joshua Podlevsky | 2012-02-01 | Mutation research |
| 21816660 | Single-stranded DNA repeat synthesis by telomerase. | Kathleen Collins | 2011-10-01 | Current opinion in chemical biology |
| 27768860 | Means to the ends: The role of telomeres and telomere processing machinery in metastasis. | J Nathaniel Robinson | 2016-12-01 | Biochimica et biophysica acta |
| 28375735 | Single-Molecule Studies of Telomeres and Telomerase. | W Joseph Parks | 2017-05-22 | Annual review of biophysics |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| TERT-204 | ENST00000484238 | 2422 | No protein | - |
| TERT-206 | ENST00000656021 | 5289 | 574aa | ENSP00000499759 |
| TERT-201 | ENST00000310581 | 4039 | 1132aa | ENSP00000309572 |
| TERT-202 | ENST00000334602 | 3210 | 1069aa | ENSP00000334346 |
| TERT-203 | ENST00000460137 | 2992 | 795aa | ENSP00000425003 |
| TERT-207 | ENST00000667927 | 493 | No protein | - |
| TERT-205 | ENST00000503656 | 406 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 16434 | Pancreatic Neoplasms | 1.046E-06 | 22523087 |
| 56341 | Glioma | 4.2800000E-009 | - |
| 56342 | Glioma | 2.1630000E-008 | - |
| 56741 | Lung Neoplasms | 4.2400000E-027 | - |
| 56778 | Adenocarcinoma of lung | 2.6750000E-025 | - |
| 57013 | Lung Diseases, Interstitial | 7.5983770E-014 | - |
| 57014 | Lung Diseases, Interstitial | 8.9298720E-007 | - |
| 60685 | Erythrocyte Indices | 1E-8 | 28017375 |
| 61774 | Leukemia, Lymphocytic, Chronic, B-Cell | 6E-8 | 26956414 |
| 63983 | Lung Diseases, Interstitial | 2E-19 | 23583980 |
| 64075 | Glioma | 4E-14 | 19578367 |
| 64599 | Melanoma | 7E-12 | 28212542 |
| 65728 | Pancreatic Carcinoma | 2E-8 | 27579533 |
| 67029 | Breast Neoplasms | 3E-14 | 27117709 |
| 67341 | Glioma | 4E-20 | 26424050 |
| 68343 | Breast Neoplasms | 3E-14 | 27117709 |
| 69229 | Breast Neoplasms | 5E-12 | 23535733 |
| 69254 | Adenocarcinoma of lung | 2E-31 | 27501781 |
| 70402 | Triple Negative Breast Neoplasms | 1E-7 | 24325915 |
| 71383 | Lung Neoplasms | 1E-27 | 21725308 |
| 73602 | Lung Neoplasms | 4E-27 | 23143601 |
| 74121 | Pancreatic Carcinoma | 1E-13 | 25086665 |
| 74864 | Erythrocyte Count | 3E-6 | 28017375 |
| 76217 | Glioma | 1E-14 | 21531791 |
| 76629 | Prostatic Neoplasms | 3E-24 | 21743467 |
| 76831 | Ovarian Neoplasms | 9E-9 | 25581431 |
| 77307 | Breast Neoplasms | 3E-8 | 27117709 |
| 78062 | Idiopathic Pulmonary Fibrosis | 3E-8 | 18835860 |
| 79373 | Adenocarcinoma of lung | 3E-11 | 20871597 |
| 80028 | Breast Neoplasms | 2E-10 | 28171663 |
| 81692 | Breast Neoplasms | 5E-6 | 28171663 |
| 82448 | Breast Neoplasms | 2E-6 | 27117709 |
| 82959 | Glioma | 1E-15 | 24908248 |
| 83768 | Thyroid Neoplasms | 3E-7 | 28195142 |
| 84220 | Glioma | 8E-7 | 26424050 |
| 85887 | Erythrocyte Count | 2E-11 | 28017375 |
| 86938 | Adenocarcinoma of lung | 2E-10 | 27197191 |
| 86939 | Breast Neoplasms | 2E-10 | 27197191 |
| 86940 | Carcinoma, Non-Small-Cell Lung | 2E-10 | 27197191 |
| 86941 | Lung Neoplasms | 2E-10 | 27197191 |
| 86942 | Ovarian Neoplasms | 2E-10 | 27197191 |
| 86943 | Prostatic Neoplasms | 2E-10 | 27197191 |
| 86944 | Colorectal Neoplasms | 2E-10 | 27197191 |
| 86945 | Carcinoma, Endometrioid | 2E-10 | 27197191 |
| 87389 | Testicular Neoplasms | 5E-24 | 23666240 |
| 88937 | Telomere | 4E-6 | 24465473 |
| 89637 | Prostatic Neoplasms | 3E-11 | 25939597 |
| 89910 | Adenocarcinoma of lung | 3E-40 | 22797724 |
| 91098 | Breast Neoplasms | 1E-8 | 27117709 |
| 91266 | Adenocarcinoma of lung | 2E-10 | 19836008 |
| 92687 | Breast Neoplasms | 7E-9 | 23535729 |
| 92923 | Glioma | 7E-9 | 21827660 |
| 93077 | Adenocarcinoma of lung | 1E-8 | 27393504 |
| 94648 | Leukemia, Lymphocytic, Chronic, B-Cell | 6E-10 | 28165464 |
| 95114 | Lung Neoplasms | 3E-9 | 27393504 |
| 95469 | Breast Neoplasms | 1E-10 | 22037553 |
| 96404 | Breast Neoplasms | 8E-28 | 27117709 |
| 97607 | Glioma | 2E-17 | 19578367 |
| 98297 | Glioma | 4E-9 | 22886559 |
| 98368 | Glioblastoma | 6E-24 | 26424050 |
| 98511 | Testicular Neoplasms | 8E-15 | 20543847 |
| 98608 | Erythrocyte Count | 3E-8 | 20139978 |
| 105321 | Adenocarcinoma of lung | 2E-22 | 20700438 |
| 106257 | Testicular Neoplasms | 1E-23 | 20543847 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
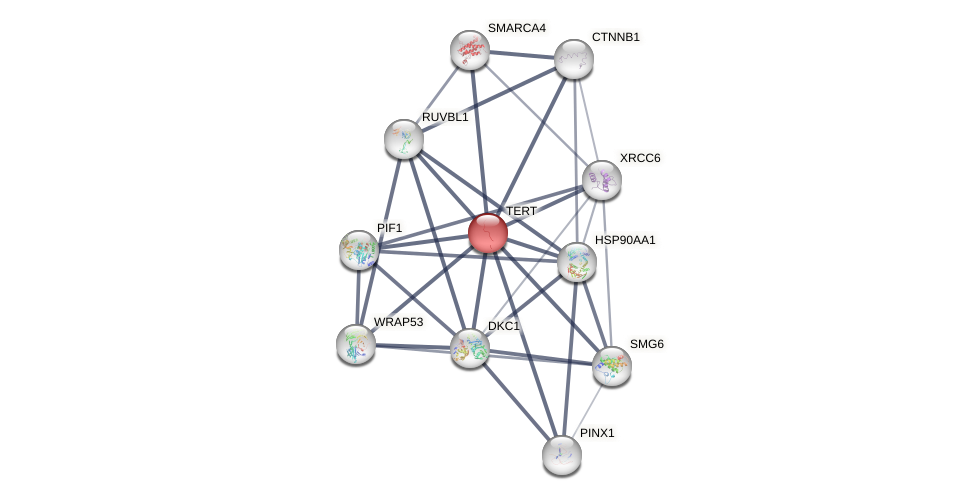
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000164362 | ||||||||
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 81.8905 | 80.7421 | nomascus_leucogenys | ENSNLEP00000013512 | 94.3032 | 92.9807 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 91.6078 | 90.9894 | pan_paniscus | ENSPPAP00000012781 | 99.7115 | 99.0385 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.5866 | 47.5265 | pongo_abelii | ENSPPYP00000025235 | 95.8188 | 93.7282 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 98.9399 | 98.4099 | pongo_abelii | ENSPPYP00000017108 | 98.9399 | 98.4099 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 67.2262 | 67.1378 | pan_troglodytes | ENSPTRP00000028637 | 99.8688 | 99.7375 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 31.6254 | 31.0954 | pan_troglodytes | ENSPTRP00000061777 | 99.4444 | 97.7778 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 99.6466 | 99.2049 | gorilla_gorilla | ENSGGOP00000002570 | 99.6466 | 99.2049 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 67.8445 | 54.3286 | ornithorhynchus_anatinus | ENSOANP00000050300 | 59.2593 | 47.4537 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 69.6996 | 54.5053 | sarcophilus_harrisii | ENSSHAP00000030586 | 58.2718 | 45.5687 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 39.2226 | 32.9505 | notamacropus_eugenii | ENSMEUP00000001231 | 43.8735 | 36.8577 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 75.4417 | 66.7845 | dasypus_novemcinctus | ENSDNOP00000031542 | 72.8669 | 64.5051 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 40.5477 | 35.2473 | echinops_telfairi | ENSETEP00000013693 | 75.7426 | 65.8416 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 90.9894 | 86.3958 | callithrix_jacchus | ENSCJAP00000070489 | 91.8822 | 87.2435 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 93.6396 | 91.2544 | cercocebus_atys | ENSCATP00000026361 | 94.39 | 91.9858 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 89.576 | 87.6325 | macaca_fascicularis | ENSMFAP00000020363 | 94.0631 | 92.0223 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 97.2615 | 95.6714 | macaca_mulatta | ENSMMUP00000014349 | 97.4336 | 95.8407 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 97.5265 | 95.8481 | macaca_nemestrina | ENSMNEP00000036262 | 97.5265 | 95.8481 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 90.5477 | 87.8975 | papio_anubis | ENSPANP00000037459 | 89.9123 | 87.2807 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 31.2721 | 30.6537 | mandrillus_leucophaeus | ENSMLEP00000015133 | 98.3333 | 96.3889 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 70.2297 | 62.5442 | tursiops_truncatus | ENSTTRP00000003636 | 70.7295 | 62.9893 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 75.7951 | 67.9329 | vulpes_vulpes | ENSVVUP00000030540 | 78.1421 | 70.0364 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 72.7915 | 62.1908 | mus_spretus | MGP_SPRETEiJ_P0036532 | 73.2444 | 62.5778 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 84.1873 | 76.6784 | equus_asinus | ENSEASP00005027225 | 84.411 | 76.8822 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 77.5618 | 69.8763 | capra_hircus | ENSCHIP00000003999 | 78.1834 | 70.4363 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 77.9152 | 69.523 | loxodonta_africana | ENSLAFP00000010943 | 80.6953 | 72.0037 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 79.3286 | 71.5548 | panthera_leo | ENSPLOP00000005238 | 80.8281 | 72.9073 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 80.5654 | 73.8516 | ailuropoda_melanoleuca | ENSAMEP00000011176 | 81.3559 | 74.5763 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 77.6502 | 69.8763 | bos_indicus_hybrid | ENSBIXP00005043343 | 79.4756 | 71.519 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 84.0106 | 76.4134 | equus_caballus | ENSECAP00000001280 | 82.6238 | 75.152 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 79.417 | 71.8198 | canis_lupus_familiaris | ENSCAFP00845036111 | 77.701 | 70.2679 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 79.2403 | 71.3781 | panthera_pardus | ENSPPRP00000013113 | 80.7381 | 72.7273 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 68.2862 | 59.8057 | marmota_marmota_marmota | ENSMMMP00000018418 | 71.1132 | 62.2815 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 81.0071 | 72.9682 | balaenoptera_musculus | ENSBMSP00010019133 | 81.3665 | 73.2919 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 71.6431 | 62.9859 | cavia_porcellus | ENSCPOP00000014030 | 79.666 | 70.0393 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 76.4134 | 68.7279 | felis_catus | ENSFCAP00000060790 | 66.4362 | 59.7542 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 80.8304 | 73.6749 | ursus_americanus | ENSUAMP00000017623 | 81.9158 | 74.6643 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 67.7562 | 52.2085 | monodelphis_domestica | ENSMODP00000036990 | 60.2041 | 46.3893 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 80.5654 | 72.2615 | monodon_monoceros | ENSMMNP00015006921 | 80.9228 | 72.5821 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 79.947 | 71.9081 | sus_scrofa | ENSSSCP00000018135 | 80.0885 | 72.0354 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 80.8304 | 73.5866 | microcebus_murinus | ENSMICP00000009921 | 83.2575 | 75.7962 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 71.5548 | 61.4841 | octodon_degus | ENSODEP00000024980 | 78.4884 | 67.4419 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 65.106 | 54.0636 | jaculus_jaculus | ENSJJAP00000011931 | 72.6108 | 60.2956 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 77.8269 | 70.4064 | ovis_aries_rambouillet | ENSOARP00020013559 | 78.4506 | 70.9706 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 54.5053 | 47.7032 | ochotona_princeps | ENSOPRP00000005699 | 64.204 | 56.1915 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 72.3498 | 64.3993 | delphinapterus_leucas | ENSDLEP00000004927 | 68.0798 | 60.5985 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 80.5654 | 72.4382 | physeter_catodon | ENSPCTP00005002003 | 81.3559 | 73.149 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 72.9682 | 62.2792 | mus_caroli | MGP_CAROLIEiJ_P0034861 | 73.4222 | 62.6667 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 72.6148 | 61.9258 | mus_musculus | ENSMUSP00000022104 | 73.262 | 62.4777 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 79.0636 | 71.0247 | mustela_putorius_furo | ENSMPUP00000008847 | 80.4133 | 72.2372 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 89.1343 | 84.5406 | aotus_nancymaae | ENSANAP00000028617 | 92.1461 | 87.3973 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 87.1025 | 82.5088 | saimiri_boliviensis_boliviensis | ENSSBOP00000028355 | 89.9635 | 85.219 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 97.2615 | 95.583 | chlorocebus_sabaeus | ENSCSAP00000013403 | 97.2615 | 95.583 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 72.1731 | 61.2191 | mesocricetus_auratus | ENSMAUP00000006231 | 73.8698 | 62.6582 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 80.1237 | 72.0848 | phocoena_sinus | ENSPSNP00000003180 | 80.4791 | 72.4046 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 80.7421 | 72.9682 | camelus_dromedarius | ENSCDRP00005010312 | 80.5286 | 72.7753 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 77.1201 | 68.7279 | carlito_syrichta | ENSTSYP00000032753 | 80.9833 | 72.1707 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 41.8728 | 36.7491 | panthera_tigris_altaica | ENSPTIP00000007073 | 73.2612 | 64.2968 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 72.5265 | 62.8092 | rattus_norvegicus | ENSRNOP00000076971 | 72.9778 | 63.2 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 25.7067 | 23.8516 | prolemur_simus | ENSPSMP00000014472 | 81.2849 | 75.419 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 71.9965 | 60.424 | microtus_ochrogaster | ENSMOCP00000002664 | 71.8695 | 60.3175 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 73.5866 | 63.0742 | peromyscus_maniculatus_bairdii | ENSPEMP00000013617 | 74.2424 | 63.6364 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 56.2721 | 49.3816 | bos_mutus | ENSBMUP00000026528 | 51.7045 | 45.3734 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 73.0565 | 62.1908 | mus_spicilegus | ENSMSIP00000020023 | 73.5111 | 62.5778 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 73.9399 | 63.0742 | cricetulus_griseus_chok1gshd | ENSCGRP00001010725 | 74.1364 | 63.2418 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 66.8728 | 59.4523 | pteropus_vampyrus | ENSPVAP00000000007 | 66.9911 | 59.5575 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 78.0035 | 69.523 | myotis_lucifugus | ENSMLUP00000011412 | 80.5657 | 71.8066 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 68.6396 | 53.8869 | vombatus_ursinus | ENSVURP00010028350 | 60.2326 | 47.2868 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 78.3569 | 70.9364 | rhinolophus_ferrumequinum | ENSRFEP00010002988 | 81.0786 | 73.4004 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 73.9399 | 65.8127 | neovison_vison | ENSNVIP00000031661 | 70.8721 | 63.0821 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 78.3569 | 70.4064 | bos_taurus | ENSBTAP00000016685 | 78.8444 | 70.8444 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 27.9152 | 24.5583 | heterocephalus_glaber_female | ENSHGLP00000009520 | 76.3285 | 67.1498 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 97.0848 | 96.0247 | rhinopithecus_bieti | ENSRBIP00000016442 | 94.1731 | 93.1448 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 79.417 | 71.8198 | canis_lupus_dingo | ENSCAFP00020029355 | 80.1963 | 72.5245 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 77.9152 | 69.788 | sciurus_vulgaris | ENSSVLP00005009543 | 79.9637 | 71.6228 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 69.8763 | 54.682 | phascolarctos_cinereus | ENSPCIP00000000271 | 58.7231 | 45.954 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 56.3604 | 50.3534 | procavia_capensis | ENSPCAP00000010230 | 58.3181 | 52.1024 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 75.9717 | 68.1095 | nannospalax_galili | ENSNGAP00000000452 | 76.3088 | 68.4117 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 78.3569 | 70.4064 | bos_grunniens | ENSBGRP00000012443 | 71.1878 | 63.9647 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 64.7526 | 56.0954 | chinchilla_lanigera | ENSCLAP00000008786 | 75.8014 | 65.667 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 77.2085 | 68.7279 | cervus_hanglu_yarkandensis | ENSCHYP00000030476 | 69.3651 | 61.746 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 43.4629 | 38.3392 | catagonus_wagneri | ENSCWAP00000000317 | 83.816 | 73.9353 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 77.3852 | 68.9929 | ictidomys_tridecemlineatus | ENSSTOP00000019046 | 81.1863 | 72.3818 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 81.8021 | 74.47 | otolemur_garnettii | ENSOGAP00000017456 | 81.8745 | 74.5358 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 91.9611 | 87.6325 | cebus_imitator | ENSCCAP00000026309 | 92.8635 | 88.4924 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 78.2686 | 69.9647 | urocitellus_parryii | ENSUPAP00010014166 | 80.4723 | 71.9346 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 74.6466 | 66.5194 | moschus_moschiferus | ENSMMSP00000024766 | 76.7484 | 68.3924 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 97.0848 | 96.1131 | rhinopithecus_roxellana | ENSRROP00000016892 | 97.0848 | 96.1131 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 73.1449 | 62.8975 | mus_pahari | MGP_PahariEiJ_P0026526 | 73.6 | 63.2889 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 54.2403 | 49.8233 | propithecus_coquereli | ENSPCOP00000025432 | 82.1954 | 75.502 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 25.3534 | 23.5866 | propithecus_coquereli | ENSPCOP00000002776 | 86.9697 | 80.9091 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 19.8763 | 8.83392 | caenorhabditis_elegans | DY3.4a.3 | 40.107 | 17.8253 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 32.5088 | 17.9329 | ciona_intestinalis | ENSCINP00000018420 | 40.708 | 22.4558 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 53.3569 | 39.1343 | eptatretus_burgeri | ENSEBUP00000009928 | 47.3354 | 34.7179 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 58.0389 | 43.4629 | callorhinchus_milii | ENSCMIP00000048136 | 55.3496 | 41.449 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 37.6325 | 27.1201 | latimeria_chalumnae | ENSLACP00000007822 | 43.7372 | 31.5195 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 53.0919 | 37.2792 | lepisosteus_oculatus | ENSLOCP00000004167 | 56.9129 | 39.9621 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 47.9682 | 34.364 | clupea_harengus | ENSCHAP00000002570 | 50.1385 | 35.9187 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 46.4664 | 31.8021 | danio_rerio | ENSDARP00000124988 | 47.9053 | 32.7869 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 24.735 | 17.7562 | carassius_auratus | ENSCARP00000128837 | 45.8265 | 32.8969 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 46.1131 | 32.8622 | carassius_auratus | ENSCARP00000015741 | 51.2267 | 36.5064 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 43.9929 | 29.417 | astyanax_mexicanus | ENSAMXP00000040764 | 47.9307 | 32.05 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 45.318 | 31.2721 | ictalurus_punctatus | ENSIPUP00000033980 | 46.7639 | 32.2698 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.8516 | 35.0707 | esox_lucius | ENSELUP00000015724 | 48.9814 | 35.1639 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 46.3781 | 33.2155 | electrophorus_electricus | ENSEEEP00000035184 | 49.435 | 35.4049 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.3816 | 36.0424 | oncorhynchus_kisutch | ENSOKIP00005011342 | 43.6037 | 31.8253 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.5583 | 36.2191 | oncorhynchus_mykiss | ENSOMYP00000136120 | 47.4619 | 34.687 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 47.4382 | 34.0989 | salmo_salar | ENSSSAP00000058453 | 46.2532 | 33.2472 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.6466 | 36.3074 | salmo_trutta | ENSSTUP00000087734 | 42.5113 | 31.0893 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.2933 | 34.629 | gadus_morhua | ENSGMOP00000065635 | 50.6812 | 35.604 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.3816 | 34.894 | fundulus_heteroclitus | ENSFHEP00000020621 | 50.8182 | 35.9091 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.2933 | 34.1873 | poecilia_reticulata | ENSPREP00000027665 | 50.8197 | 35.2459 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.3816 | 34.7173 | xiphophorus_maculatus | ENSXMAP00000016432 | 50.9572 | 35.825 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.5866 | 34.0989 | oryzias_latipes | ENSORLP00000017467 | 50.4587 | 35.4128 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.8516 | 35.5124 | cyclopterus_lumpus | ENSCLMP00005014452 | 50.9208 | 37.0166 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.1449 | 33.8339 | oreochromis_niloticus | ENSONIP00000040184 | 46.7009 | 32.8192 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 47.5265 | 33.2155 | haplochromis_burtoni | ENSHBUP00000032466 | 49.631 | 34.6863 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.2332 | 33.7456 | astatotilapia_calliptera | ENSACLP00000006476 | 49.863 | 34.8858 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.4099 | 34.5406 | sparus_aurata | ENSSAUP00010010436 | 49.3694 | 35.2252 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 47.5265 | 33.8339 | lates_calcarifer | ENSLCAP00010053616 | 48.9091 | 34.8182 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 60.6007 | 44.3463 | xenopus_tropicalis | ENSXETP00000034113 | 55.6367 | 40.7137 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 65.0177 | 50.3534 | chrysemys_picta_bellii | ENSCPBP00000022371 | 54.4379 | 42.1598 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 60.2473 | 45.6714 | sphenodon_punctatus | ENSSPUP00000005438 | 61.2208 | 46.4093 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 62.0141 | 48.2332 | crocodylus_porosus | ENSCPRP00005020350 | 55.1887 | 42.9245 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 59.4523 | 43.8163 | laticauda_laticaudata | ENSLLTP00000016955 | 53.6683 | 39.5534 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 62.0141 | 45.8481 | notechis_scutatus | ENSNSUP00000013627 | 53.917 | 39.8618 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 59.4523 | 43.8163 | pseudonaja_textilis | ENSPTXP00000005667 | 53.5828 | 39.4904 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 62.9859 | 47.0848 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000017788 | 52.9324 | 39.5694 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 62.8975 | 48.3216 | gallus_gallus | ENSGALP00010010988 | 52.43 | 40.2798 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 59.364 | 45.4947 | meleagris_gallopavo | ENSMGAP00000017222 | 53.5458 | 41.0359 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 59.629 | 45.318 | serinus_canaria | ENSSCAP00000007871 | 52.4476 | 39.8601 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 62.0141 | 46.5548 | parus_major | ENSPMJP00000018341 | 46.0932 | 34.6028 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 62.8975 | 46.2014 | anolis_carolinensis | ENSACAP00000001407 | 53.3733 | 39.2054 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.2332 | 33.8339 | neolamprologus_brichardi | ENSNBRP00000026654 | 49.863 | 34.9772 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 59.0106 | 43.2862 | naja_naja | ENSNNAP00000001373 | 52.8899 | 38.7965 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 64.311 | 48.0565 | podarcis_muralis | ENSPMRP00000017796 | 51.4851 | 38.4724 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 56.5371 | 43.0212 | geospiza_fortis | ENSGFOP00000009433 | 53.2003 | 40.4821 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 62.3675 | 47.5265 | taeniopygia_guttata | ENSTGUP00000023803 | 52.5689 | 40.0596 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 58.7456 | 42.3145 | erpetoichthys_calabaricus | ENSECRP00000018670 | 53.1575 | 38.2894 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.2049 | 33.6572 | kryptolebias_marmoratus | ENSKMAP00000029334 | 50.4529 | 34.5109 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 62.6325 | 47.2615 | salvator_merianae | ENSSMRP00000019897 | 56.2252 | 42.4266 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.1449 | 33.3039 | oryzias_javanicus | ENSOJAP00000029653 | 50.138 | 34.6826 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 47.8799 | 33.7456 | amphilophus_citrinellus | ENSACIP00000022588 | 49.3625 | 34.7905 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.3216 | 34.4523 | dicentrarchus_labrax | ENSDLAP00005032084 | 49.8179 | 35.5191 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.4099 | 33.6572 | amphiprion_percula | ENSAPEP00000001559 | 49.4139 | 34.3553 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 61.2191 | 46.9081 | pelodiscus_sinensis | ENSPSIP00000014126 | 55.1752 | 42.2771 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 64.576 | 49.5583 | gopherus_evgoodei | ENSGEVP00005002876 | 55.3369 | 42.4678 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 64.0459 | 48.8516 | chelonoidis_abingdonii | ENSCABP00000008802 | 54.9242 | 41.8939 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.9117 | 35.5124 | seriola_dumerili | ENSSDUP00000002490 | 50.9928 | 36.2816 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.9399 | 34.7173 | cottoperca_gobio | ENSCGOP00000011056 | 50 | 35.4693 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 58.1272 | 43.9046 | anser_brachyrhynchus | ENSABRP00000022781 | 54.0674 | 40.8381 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.9399 | 35.159 | gasterosteus_aculeatus | ENSGACP00000012238 | 50.0452 | 35.953 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 45.318 | 31.1837 | cynoglossus_semilaevis | ENSCSEP00000015118 | 50.0977 | 34.4727 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.1449 | 33.5689 | poecilia_formosa | ENSPFOP00000015052 | 50.1842 | 34.9908 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.9117 | 34.8057 | myripristis_murdjan | ENSMMDP00005010575 | 50.8094 | 35.4317 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.7633 | 34.9823 | sander_lucioperca | ENSSLUP00000049793 | 49.8645 | 35.7724 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 62.4558 | 47.9682 | strigops_habroptila | ENSSHBP00005005413 | 53.238 | 40.8886 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.3216 | 33.6572 | amphiprion_ocellaris | ENSAOCP00000019752 | 49.3682 | 34.3863 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 65.0177 | 50.1767 | terrapene_carolina_triunguis | ENSTMTP00000011513 | 54.3575 | 41.9498 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 45.2297 | 32.4205 | hucho_hucho | ENSHHUP00000068133 | 49.8054 | 35.7004 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.3816 | 35.0707 | betta_splendens | ENSBSLP00000008574 | 49.7774 | 35.3517 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 44.4346 | 30.7421 | pygocentrus_nattereri | ENSPNAP00000003578 | 47.9048 | 33.1429 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.0283 | 34.7173 | anabas_testudineus | ENSATEP00000051929 | 50.0903 | 35.4693 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 30.7421 | 16.3428 | saccharomyces_cerevisiae | YLR318W | 39.3665 | 20.9276 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.735 | 35.6007 | seriola_lalandi_dorsalis | ENSSLDP00000010197 | 50.8123 | 36.3718 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.47 | 34.1873 | labrus_bergylta | ENSLBEP00000031507 | 50.5415 | 34.9278 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 47.4382 | 32.8622 | pundamilia_nyererei | ENSPNYP00000000968 | 49.2661 | 34.1284 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.2049 | 34.4523 | mastacembelus_armatus | ENSMAMP00000009150 | 50.1802 | 35.1351 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.0283 | 34.4523 | stegastes_partitus | ENSSPAP00000027196 | 50.1355 | 35.2304 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.1166 | 33.8339 | acanthochromis_polyacanthus | ENSAPOP00000017292 | 50.1353 | 34.5356 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 63.1625 | 48.3216 | aquila_chrysaetos_chrysaetos | ENSACCP00020003525 | 53.7999 | 41.1588 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.2049 | 34.4523 | cyprinodon_variegatus | ENSCVAP00000028456 | 51.0541 | 35.747 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 46.3781 | 31.9788 | denticeps_clupeoides | ENSDCDP00000042703 | 50.5294 | 34.8412 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.3816 | 35.0707 | larimichthys_crocea | ENSLCRP00005032933 | 50.5425 | 35.8951 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.4982 | 33.4806 | takifugu_rubripes | ENSTRUP00000036305 | 51.1173 | 35.2886 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 58.6572 | 44.523 | ficedula_albicollis | ENSFALP00000027920 | 53.1625 | 40.3523 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 45.6714 | 31.8905 | hippocampus_comes | ENSHCOP00000005022 | 48.5446 | 33.8967 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 46.7314 | 33.7456 | cyprinus_carpio_carpio | ENSCCRP00000129796 | 49.5783 | 35.8013 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 47.0848 | 33.6572 | cyprinus_carpio_carpio | ENSCCRP00000179009 | 51.0048 | 36.4593 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 62.7208 | 48.2332 | coturnix_japonica | ENSCJPP00005018191 | 52.8274 | 40.625 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.1166 | 34.2756 | poecilia_latipinna | ENSPLAP00000015896 | 50.6837 | 35.3692 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 48.2332 | 33.7456 | maylandia_zebra | ENSMZEP00005015090 | 49.863 | 34.8858 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 47.2615 | 32.6855 | tetraodon_nigroviridis | ENSTNIP00000001073 | 49.2634 | 34.07 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.2049 | 35.8657 | paramormyrops_kingsleyae | ENSPKIP00000005810 | 52.9468 | 38.5932 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 46.9965 | 32.4205 | scleropages_formosus | ENSSFOP00015019231 | 51.401 | 35.4589 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 57.9505 | 42.3145 | leptobrachium_leishanense | ENSLLEP00000041193 | 52.7331 | 38.5048 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 49.1166 | 34.9823 | scophthalmus_maximus | ENSSMAP00000033907 | 51.3863 | 36.5989 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 47.4382 | 32.0671 | oryzias_melastigma | ENSOMEP00000011516 | 49.4475 | 33.4254 |
| ENSG00000164362 | homo_sapiens | ENSP00000309572 | 37.4558 | 25.265 | oryzias_melastigma | ENSOMEP00000005848 | 52.4104 | 35.3523 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000049 | enables tRNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0000333 | part_of telomerase catalytic core complex | 4 | IBA,IDA,IMP,IPI | Homo_sapiens(9606) | Component |
| GO:0000723 | involved_in telomere maintenance | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0000781 | located_in chromosome, telomeric region | 2 | IC,IDA | Homo_sapiens(9606) | Component |
| GO:0000783 | part_of nuclear telomere cap complex | 1 | IC | Homo_sapiens(9606) | Component |
| GO:0001172 | involved_in RNA-templated transcription | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0001223 | enables transcription coactivator binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0003677 | enables DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003720 | enables telomerase activity | 2 | IDA,TAS | Homo_sapiens(9606) | Function |
| GO:0003721 | enables telomerase RNA reverse transcriptase activity | 4 | IBA,IDA,IMP,TAS | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0003964 | enables RNA-directed DNA polymerase activity | 2 | IDA,TAS | Homo_sapiens(9606) | Function |
| GO:0003968 | enables RNA-dependent RNA polymerase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005697 | part_of telomerase holoenzyme complex | 4 | IDA,IPI,NAS,TAS | Homo_sapiens(9606) | Component |
| GO:0005730 | located_in nucleolus | 2 | IDA,NAS | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005886 | located_in plasma membrane | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0006278 | involved_in RNA-templated DNA biosynthetic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0007004 | involved_in telomere maintenance via telomerase | 4 | IBA,IDA,IMP,NAS | Homo_sapiens(9606) | Process |
| GO:0007005 | involved_in mitochondrion organization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0010629 | involved_in negative regulation of gene expression | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0016605 | colocalizes_with PML body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0016607 | located_in nuclear speck | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0022616 | involved_in DNA strand elongation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0030177 | involved_in positive regulation of Wnt signaling pathway | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:0030422 | involved_in siRNA processing | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0031379 | part_of RNA-directed RNA polymerase complex | 1 | IPI | Homo_sapiens(9606) | Component |
| GO:0031647 | involved_in regulation of protein stability | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0032092 | involved_in positive regulation of protein binding | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0042162 | enables telomeric DNA binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0042635 | involved_in positive regulation of hair cycle | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0042645 | located_in mitochondrial nucleoid | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0042802 | enables identical protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0042803 | enables protein homodimerization activity | 2 | IDA,IPI | Homo_sapiens(9606) | Function |
| GO:0043524 | involved_in negative regulation of neuron apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045766 | involved_in positive regulation of angiogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0046326 | involved_in positive regulation of glucose import | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0046686 | involved_in response to cadmium ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0046872 | enables metal ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0051000 | involved_in positive regulation of nitric-oxide synthase activity | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051087 | enables protein-folding chaperone binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0070034 | enables telomerase RNA binding | 3 | IBA,IDA,IPI | Homo_sapiens(9606) | Function |
| GO:0070200 | involved_in establishment of protein localization to telomere | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071456 | involved_in cellular response to hypoxia | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0071897 | involved_in DNA biosynthetic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0090399 | involved_in replicative senescence | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0098680 | enables template-free RNA nucleotidyltransferase | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0140745 | involved_in siRNA transcription | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1900087 | involved_in positive regulation of G1/S transition of mitotic cell cycle | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1902895 | involved_in positive regulation of miRNA transcription | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1903620 | involved_in positive regulation of transdifferentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1904707 | involved_in positive regulation of vascular associated smooth muscle cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1904751 | involved_in positive regulation of protein localization to nucleolus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1904754 | involved_in positive regulation of vascular associated smooth muscle cell migration | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1990572 | part_of TERT-RMRP complex | 2 | IDA,IPI | Homo_sapiens(9606) | Component |
| GO:2000352 | involved_in negative regulation of endothelial cell apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:2000648 | involved_in positive regulation of stem cell proliferation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:2000773 | involved_in negative regulation of cellular senescence | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:2001240 | involved_in negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 1 | IMP | Homo_sapiens(9606) | Process |