Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000148090 | Gene ID | 549 | Accession | 890 |
| Symbol | AUH | Alias | NA | Full Name | AU RNA binding methylglutaconyl-CoA hydratase |
| Status | Confidence | Length | 148104 bases | Strand | Minus strand |
| Position | 9 : 91213815 - 91361918 | RNA binding domain | N.A. | ||
| Summary | This gene encodes bifunctional mitochondrial protein that has both RNA-binding and hydratase activities. The encoded protein is a methylglutaconyl-CoA hydratase that catalyzes the hydration of 3-methylglutaconyl-CoA to 3-hydroxy-3-methyl-glutaryl-CoA, a critical step in the leucine degradation pathway. This protein also binds AU-rich elements (AREs) found in the 3' UTRs of rapidly decaying mRNAs including c-fos, c-myc and granulocyte/ macrophage colony stimulating factor. ARE elements are involved in directing RNA to rapid degradation and deadenylation. This protein is localizes to the mitochondrial matrix and the inner mitochondrial membrane and may be involved in mitochondrial protein synthesis. Mutations in this gene are the cause of 3-methylglutaconic aciduria, type I. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Sep 2015] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 18221507 | Development of a human mitochondrial oligonucleotide microarray (h-MitoArray) and gene expression analysis of fibroblast cell lines from 13 patients with isolated F1Fo ATP synthase deficiency. | Alena Cízková | 2008-01-25 | BMC genomics |
| 24598254 | A bifunctional protein regulates mitochondrial protein synthesis. | R Tara Richman | 2014-05-01 | Nucleic acids research |
| 7892223 | AUH, a gene encoding an AU-specific RNA binding protein with intrinsic enoyl-CoA hydratase activity. | J Nakagawa | 1995-03-14 | Proceedings of the National Academy of Sciences of the United States of America |
| 25695250 | Whole cell formaldehyde cross-linking simplifies purification of mitochondrial nucleoids and associated proteins involved in mitochondrial gene expression. | Nina Rajala | 2015-01-01 | PloS one |
| 31106228 | De Novo Interstitial Deletion of 9q in a Pediatric Patient With Global Developmental Delay. | Dennis Keselman | 2019-01-01 | Child neurology open |
| 24989451 | EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. | Veronika Boczonadi | 2014-07-03 | Nature communications |
| 22904065 | p32/gC1qR is indispensable for fetal development and mitochondrial translation: importance of its RNA-binding ability. | Mikako Yagi | 2012-10-01 | Nucleic acids research |
| 34221536 | Large-scale analysis of the position-dependent binding and regulation of human RNA binding proteins. | Jianan Lin | 2020-06-01 | Quantitative biology (Beijing, China) |
| 33193737 | KPNA2-Associated Immune Analyses Highlight the Dysregulation and Prognostic Effects of GRB2, NRAS, and Their RNA-Binding Proteins in Hepatocellular Carcinoma. | Xiuzhi Zhang | 2020-01-01 | Frontiers in genetics |
| 26189680 | A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. | Oren Parnas | 2015-07-30 | Cell |
| 11738050 | Crystal structure of human AUH protein, a single-stranded RNA binding homolog of enoyl-CoA hydratase. | K Kurimoto | 2001-12-01 | Structure (London, England : 1993) |
| 19196430 | Mutant copper-zinc superoxide dismutase associated with amyotrophic lateral sclerosis binds to adenine/uridine-rich stability elements in the vascular endothelial growth factor 3'-untranslated region. | Xuelin Li | 2009-02-01 | Journal of neurochemistry |
| 21909410 | Novel susceptibility locus at 22q11 for diabetic nephropathy in type 1 diabetes. | Maija Wessman | 2011-01-01 | PloS one |
| 16640564 | Biochemical characterization of human 3-methylglutaconyl-CoA hydratase and its role in leucine metabolism. | Matthias Mack | 2006-05-01 | The FEBS journal |
| 12434311 | 3-Methylglutaconic aciduria type I is caused by mutations in AUH. | Lodewijk IJlst | 2002-12-01 | American journal of human genetics |
| 11343630 | Mood stabilizers regulate cytoprotective and mRNA-binding proteins in the brain: long-term effects on cell survival and transcript stability. | G Chen | 2001-03-01 | The international journal of neuropsychopharmacology |
| 21150874 | A genome-wide association study for diabetic nephropathy genes in African Americans. | W Caitrin McDonough | 2011-03-01 | Kidney international |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| AUH-202 | ENST00000375731 | 1574 | 339aa | ENSP00000364883 |
| AUH-201 | ENST00000303617 | 1490 | 310aa | ENSP00000307334 |
| AUH-203 | ENST00000473695 | 632 | No protein | - |
| AUH-205 | ENST00000478465 | 751 | No protein | - |
| AUH-204 | ENST00000475023 | 641 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 422 | Respiratory Function Tests | 1.22718160066294E-7 | 17903307 |
| 8264 | Electrocardiography | 2.65392231829385E-10 | 17903306 |
| 13307 | Lupus Erythematosus, Systemic | 2.37E-6 | 18204446 |
| 14987 | Myocardial Infarction | 5.3662451E-004 | - |
| 15154 | Myocardial Infarction | 8.0106382E-004 | - |
| 29903 | Triglycerides | 9.5590000E-005 | - |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
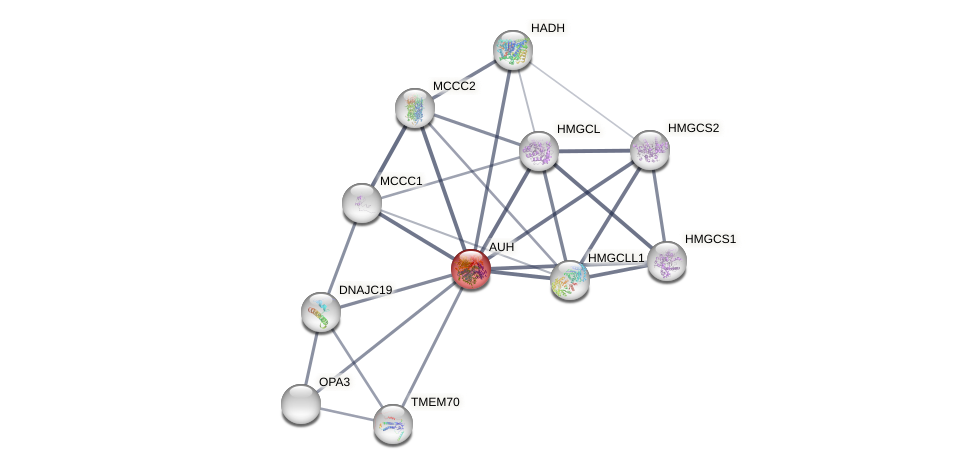
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000148090 | homo_sapiens | ENSP00000401751 | 44.5183 | 27.2425 | homo_sapiens | ENSP00000364883 | 39.528 | 24.1888 |
| ENSG00000148090 | homo_sapiens | ENSP00000476295 | 24.7036 | 13.4387 | homo_sapiens | ENSP00000364883 | 36.8732 | 20.059 |
| ENSG00000148090 | homo_sapiens | ENSP00000301729 | 39.7351 | 22.8477 | homo_sapiens | ENSP00000364883 | 35.3982 | 20.354 |
| ENSG00000148090 | homo_sapiens | ENSP00000352706 | 34.715 | 19.4301 | homo_sapiens | ENSP00000364883 | 39.528 | 22.1239 |
| ENSG00000148090 | homo_sapiens | ENSP00000368517 | 42.2442 | 24.7525 | homo_sapiens | ENSP00000364883 | 37.7581 | 22.1239 |
| ENSG00000148090 | homo_sapiens | ENSP00000302968 | 21.6607 | 11.1913 | homo_sapiens | ENSP00000364883 | 35.3982 | 18.2891 |
| ENSG00000148090 | homo_sapiens | ENSP00000250838 | 22.7357 | 11.8299 | homo_sapiens | ENSP00000364883 | 36.2832 | 18.8791 |
| ENSG00000148090 | homo_sapiens | ENSP00000371844 | 22.5926 | 11.8519 | homo_sapiens | ENSP00000364883 | 35.9882 | 18.8791 |
| ENSG00000148090 | homo_sapiens | ENSP00000372319 | 22.7357 | 11.8299 | homo_sapiens | ENSP00000364883 | 36.2832 | 18.8791 |
| ENSG00000148090 | homo_sapiens | ENSP00000380718 | 22.9779 | 13.0515 | homo_sapiens | ENSP00000364883 | 36.8732 | 20.944 |
| ENSG00000148090 | homo_sapiens | ENSP00000221418 | 42.0732 | 25 | homo_sapiens | ENSP00000364883 | 40.708 | 24.1888 |
| ENSG00000148090 | homo_sapiens | ENSP00000360577 | 72.2603 | 52.3973 | homo_sapiens | ENSP00000364883 | 62.2419 | 45.1327 |
| ENSG00000148090 | homo_sapiens | ENSP00000357535 | 51.0345 | 33.1034 | homo_sapiens | ENSP00000364883 | 43.6578 | 28.3186 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 75.2212 | 71.0914 | nomascus_leucogenys | ENSNLEP00000034918 | 81.4697 | 76.9968 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 76.1062 | 74.6313 | pan_paniscus | ENSPPAP00000031480 | 64.8241 | 63.5678 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 98.8201 | 98.5251 | pongo_abelii | ENSPPYP00000021702 | 98.8201 | 98.5251 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 99.705 | 99.115 | pan_troglodytes | ENSPTRP00000079432 | 99.705 | 99.115 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 99.705 | 99.41 | gorilla_gorilla | ENSGGOP00000003756 | 99.705 | 99.41 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 75.8112 | 64.6018 | ornithorhynchus_anatinus | ENSOANP00000036804 | 58.0135 | 49.4357 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 82.3009 | 73.4513 | sarcophilus_harrisii | ENSSHAP00000040885 | 84.0361 | 75 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 82.5959 | 76.1062 | notamacropus_eugenii | ENSMEUP00000004179 | 85.6269 | 78.8991 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 81.7109 | 78.1711 | choloepus_hoffmanni | ENSCHOP00000012397 | 81.7109 | 78.1711 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 57.8171 | 55.4572 | dasypus_novemcinctus | ENSDNOP00000034547 | 97.5124 | 93.5323 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 87.6106 | 83.7758 | erinaceus_europaeus | ENSEEUP00000010265 | 88.3929 | 84.5238 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 97.9351 | 96.1652 | callithrix_jacchus | ENSCJAP00000010488 | 97.9351 | 96.1652 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.9705 | 87.0207 | callithrix_jacchus | ENSCJAP00000089264 | 92.4242 | 89.3939 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 95.2802 | 94.3953 | cercocebus_atys | ENSCATP00000037999 | 97.5831 | 96.6767 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 99.115 | 98.2301 | macaca_fascicularis | ENSMFAP00000051155 | 88.8889 | 88.0952 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 98.8201 | 97.9351 | macaca_mulatta | ENSMMUP00000020756 | 88.6243 | 87.8307 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 99.115 | 98.2301 | macaca_nemestrina | ENSMNEP00000042683 | 99.115 | 98.2301 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 98.8201 | 98.2301 | papio_anubis | ENSPANP00000019399 | 98.8201 | 98.2301 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 66.9617 | 65.1917 | mandrillus_leucophaeus | ENSMLEP00000032773 | 91.9028 | 89.4737 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 67.2566 | 64.8968 | tursiops_truncatus | ENSTTRP00000011785 | 67.8571 | 65.4762 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 93.2153 | 88.7906 | vulpes_vulpes | ENSVVUP00000030588 | 92.6686 | 88.2698 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.3805 | 87.6106 | mus_spretus | MGP_SPRETEiJ_P0036000 | 96.4968 | 94.586 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 93.8053 | 89.9705 | equus_asinus | ENSEASP00005035976 | 94.9254 | 91.0448 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 92.6254 | 89.0855 | capra_hircus | ENSCHIP00000016884 | 92.6254 | 89.0855 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 84.0708 | 78.4661 | loxodonta_africana | ENSLAFP00000014279 | 91.0543 | 84.984 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 90.2655 | 84.9557 | panthera_leo | ENSPLOP00000015623 | 75 | 70.5882 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 93.5103 | 90.8555 | ailuropoda_melanoleuca | ENSAMEP00000035255 | 93.5103 | 90.8555 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.6814 | 67.5516 | bos_indicus_hybrid | ENSBIXP00005045481 | 92.7481 | 87.4046 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 92.9204 | 89.3805 | bos_indicus_hybrid | ENSBIXP00005010930 | 92.9204 | 89.3805 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 77.8761 | 75.2212 | oryctolagus_cuniculus | ENSOCUP00000006425 | 78.3383 | 75.6677 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 81.7109 | 74.3363 | equus_caballus | ENSECAP00000073616 | 82.4405 | 75 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 93.8053 | 89.0855 | canis_lupus_familiaris | ENSCAFP00845024861 | 93.5294 | 88.8235 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 93.5103 | 89.9705 | panthera_pardus | ENSPPRP00000002129 | 93.5103 | 89.9705 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 32.4484 | 30.9735 | marmota_marmota_marmota | ENSMMMP00000025052 | 88 | 84 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 92.6254 | 89.3805 | balaenoptera_musculus | ENSBMSP00010006995 | 92.6254 | 89.3805 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 74.6313 | 72.8614 | cavia_porcellus | ENSCPOP00000000963 | 96.1977 | 93.9164 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 91.1504 | 85.5457 | felis_catus | ENSFCAP00000060298 | 62.5506 | 58.7045 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.3805 | 86.4307 | ursus_americanus | ENSUAMP00000032001 | 91.5408 | 88.5196 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 77.2861 | 71.0914 | monodelphis_domestica | ENSMODP00000057758 | 63.9024 | 58.7805 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 68.4366 | 64.3068 | bison_bison_bison | ENSBBBP00000003921 | 86.5672 | 81.3433 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.3805 | 86.4307 | bison_bison_bison | ENSBBBP00000002856 | 89.3805 | 86.4307 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.6755 | 87.3156 | monodon_monoceros | ENSMMNP00015030214 | 89.6755 | 87.3156 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 91.4454 | 89.0855 | monodon_monoceros | ENSMMNP00015016468 | 90.379 | 88.0466 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 92.0354 | 88.4956 | sus_scrofa | ENSSSCP00000010238 | 92.3077 | 88.7574 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 95.8702 | 94.3953 | microcebus_murinus | ENSMICP00000040577 | 95.8702 | 94.3953 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 80.826 | 75.2212 | octodon_degus | ENSODEP00000001181 | 84.0491 | 78.2209 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.0855 | 87.3156 | jaculus_jaculus | ENSJJAP00000011740 | 92.0732 | 90.2439 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 92.9204 | 89.3805 | ovis_aries_rambouillet | ENSOARP00020000503 | 92.9204 | 89.3805 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 73.1563 | 70.5015 | ursus_maritimus | ENSUMAP00000022632 | 79.7428 | 76.8489 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 70.7965 | 68.7316 | ochotona_princeps | ENSOPRP00000000063 | 76.9231 | 74.6795 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 68.4366 | 66.6667 | delphinapterus_leucas | ENSDLEP00000026499 | 88.5496 | 86.2595 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 87.0207 | 84.6608 | delphinapterus_leucas | ENSDLEP00000025446 | 89.6657 | 87.234 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 92.0354 | 89.0855 | physeter_catodon | ENSPCTP00005015282 | 92.0354 | 89.0855 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.3805 | 87.3156 | mus_caroli | MGP_CAROLIEiJ_P0034367 | 96.4968 | 94.2675 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.3805 | 87.3156 | mus_musculus | ENSMUSP00000021913 | 96.4968 | 94.2675 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 85.5457 | 83.1858 | dipodomys_ordii | ENSDORP00000017267 | 91.195 | 88.6792 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 66.3717 | 64.3068 | mustela_putorius_furo | ENSMPUP00000006792 | 97.8261 | 94.7826 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 97.9351 | 96.4602 | aotus_nancymaae | ENSANAP00000007202 | 97.9351 | 96.4602 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 97.3451 | 95.8702 | saimiri_boliviensis_boliviensis | ENSSBOP00000012545 | 97.3451 | 95.8702 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 61.0619 | 59.882 | vicugna_pacos | ENSVPAP00000000148 | 82.4701 | 80.8765 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 98.5251 | 97.9351 | chlorocebus_sabaeus | ENSCSAP00000003541 | 98.2353 | 97.6471 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 50.1475 | 48.6726 | tupaia_belangeri | ENSTBEP00000008023 | 72.3404 | 70.2128 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.3805 | 86.7257 | mesocricetus_auratus | ENSMAUP00000008034 | 96.4968 | 93.6306 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 72.2714 | 68.7316 | phocoena_sinus | ENSPSNP00000007746 | 87.8136 | 83.5125 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 91.7404 | 89.3805 | phocoena_sinus | ENSPSNP00000001309 | 91.7404 | 89.3805 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 53.6873 | 52.2124 | camelus_dromedarius | ENSCDRP00005024850 | 93.8144 | 91.2371 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 72.8614 | 70.7965 | carlito_syrichta | ENSTSYP00000003775 | 72.8614 | 70.7965 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.9764 | 69.9115 | panthera_tigris_altaica | ENSPTIP00000007845 | 71.9764 | 69.9115 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.0855 | 87.3156 | rattus_norvegicus | ENSRNOP00000015786 | 95.873 | 93.9683 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 94.9853 | 92.3304 | prolemur_simus | ENSPSMP00000036839 | 94.9853 | 92.3304 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.3805 | 87.9056 | microtus_ochrogaster | ENSMOCP00000014114 | 96.4968 | 94.9045 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 55.1622 | 52.8024 | sorex_araneus | ENSSARP00000002615 | 74.502 | 71.3147 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.9705 | 88.2006 | peromyscus_maniculatus_bairdii | ENSPEMP00000012318 | 97.1338 | 95.2229 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 38.3481 | 35.1032 | bos_mutus | ENSBMUP00000009000 | 87.8378 | 80.4054 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 76.6962 | 74.6313 | mus_spicilegus | ENSMSIP00000008242 | 84.1424 | 81.877 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 88.7906 | 86.7257 | cricetulus_griseus_chok1gshd | ENSCGRP00001014719 | 95.2532 | 93.038 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 90.2655 | 85.8407 | pteropus_vampyrus | ENSPVAP00000004541 | 90.8012 | 86.3502 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 14.7493 | 13.5693 | myotis_lucifugus | ENSMLUP00000002869 | 96.1538 | 88.4615 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 82.3009 | 76.1062 | vombatus_ursinus | ENSVURP00010008871 | 84.0361 | 77.7108 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 91.7404 | 87.0207 | rhinolophus_ferrumequinum | ENSRFEP00010018550 | 92.2849 | 87.5371 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 74.0413 | 71.0914 | neovison_vison | ENSNVIP00000020046 | 88.3803 | 84.8592 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 37.7581 | 31.8584 | bos_taurus | ENSBTAP00000068994 | 66.3212 | 55.9585 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 73.7463 | 70.2065 | bos_taurus | ENSBTAP00000067281 | 90.9091 | 86.5455 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 40.118 | 37.1681 | heterocephalus_glaber_female | ENSHGLP00000042062 | 83.9506 | 77.7778 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 98.8201 | 98.2301 | rhinopithecus_bieti | ENSRBIP00000009555 | 99.1124 | 98.5207 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 86.7257 | 80.826 | canis_lupus_dingo | ENSCAFP00020008352 | 88.0239 | 82.0359 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 39.233 | 35.6932 | sciurus_vulgaris | ENSSVLP00005009428 | 65.5172 | 59.6059 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 75.8112 | 65.7817 | phascolarctos_cinereus | ENSPCIP00000047590 | 59.0805 | 51.2644 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 63.1268 | 60.767 | procavia_capensis | ENSPCAP00000014700 | 63.1268 | 60.767 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.3805 | 86.4307 | nannospalax_galili | ENSNGAP00000007744 | 96.4968 | 93.3121 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 36.2832 | 35.6932 | bos_grunniens | ENSBGRP00000014154 | 91.791 | 90.2985 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 81.7109 | 79.056 | chinchilla_lanigera | ENSCLAP00000000854 | 96.1806 | 93.0556 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 92.9204 | 89.0855 | cervus_hanglu_yarkandensis | ENSCHYP00000014983 | 92.9204 | 89.0855 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 92.6254 | 87.9056 | catagonus_wagneri | ENSCWAP00000001747 | 92.8994 | 88.1657 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 67.2566 | 63.7168 | ictidomys_tridecemlineatus | ENSSTOP00000024366 | 85.3933 | 80.8989 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 94.9853 | 92.0354 | otolemur_garnettii | ENSOGAP00000010512 | 95.8333 | 92.8571 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 96.7552 | 94.9853 | cebus_imitator | ENSCCAP00000002723 | 96.7552 | 94.9853 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 93.2153 | 90.2655 | moschus_moschiferus | ENSMMSP00000017877 | 93.2153 | 90.2655 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 92.6254 | 91.4454 | rhinopithecus_roxellana | ENSRROP00000031798 | 92.0821 | 90.9091 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 89.0855 | 87.3156 | mus_pahari | MGP_PahariEiJ_P0040975 | 96.1783 | 94.2675 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 97.0501 | 95.2802 | propithecus_coquereli | ENSPCOP00000019109 | 97.0501 | 95.2802 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 58.1121 | 41.5929 | caenorhabditis_elegans | F56B3.5.1 | 68.6411 | 49.1289 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 61.9469 | 48.0826 | drosophila_melanogaster | FBpp0086993 | 70.2341 | 54.515 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 60.177 | 47.1976 | ciona_intestinalis | ENSCINP00000005607 | 67.7741 | 53.1561 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 50.7375 | 37.4631 | petromyzon_marinus | ENSPMAP00000003770 | 66.9261 | 49.4163 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 64.8968 | 50.4425 | eptatretus_burgeri | ENSEBUP00000012565 | 72.3684 | 56.25 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.3864 | 58.9971 | callorhinchus_milii | ENSCMIP00000028277 | 72.6727 | 60.0601 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 59.587 | 50.1475 | latimeria_chalumnae | ENSLACP00000008049 | 71.6312 | 60.2837 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 73.4513 | 62.8319 | lepisosteus_oculatus | ENSLOCP00000009963 | 74.7748 | 63.964 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.3864 | 58.4071 | clupea_harengus | ENSCHAP00000043036 | 74.4615 | 60.9231 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 73.4513 | 58.7021 | danio_rerio | ENSDARP00000063092 | 76.6154 | 61.2308 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.6165 | 56.0472 | carassius_auratus | ENSCARP00000081056 | 73.9812 | 59.5611 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 70.7965 | 58.4071 | astyanax_mexicanus | ENSAMXP00000052532 | 76.6773 | 63.2588 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.6814 | 57.8171 | ictalurus_punctatus | ENSIPUP00000037313 | 66.0326 | 53.2609 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 63.7168 | 49.5575 | esox_lucius | ENSELUP00000037258 | 63.9053 | 49.7041 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 67.5516 | 56.9322 | electrophorus_electricus | ENSEEEP00000041889 | 66.3768 | 55.942 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.0914 | 57.5221 | oncorhynchus_kisutch | ENSOKIP00005117754 | 73.9264 | 59.816 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 63.4218 | 51.0324 | oncorhynchus_mykiss | ENSOMYP00000126669 | 42.0744 | 33.8552 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 70.7965 | 57.8171 | salmo_salar | ENSSSAP00000106453 | 68.5714 | 56 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.6814 | 58.4071 | salmo_trutta | ENSSTUP00000059212 | 67.313 | 54.8476 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.9115 | 57.2271 | gadus_morhua | ENSGMOP00000055617 | 73.6025 | 60.2484 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 34.8083 | 24.7788 | fundulus_heteroclitus | ENSFHEP00000013749 | 57.8431 | 41.1765 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 52.8024 | 41.8879 | fundulus_heteroclitus | ENSFHEP00000028945 | 66.0517 | 52.3985 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.0266 | 57.2271 | poecilia_reticulata | ENSPREP00000006396 | 73.3542 | 60.815 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.0266 | 58.7021 | xiphophorus_maculatus | ENSXMAP00000016170 | 72.8972 | 61.9938 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.6814 | 59.882 | oryzias_latipes | ENSORLP00000025083 | 75.2322 | 62.8483 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.3215 | 58.4071 | cyclopterus_lumpus | ENSCLMP00005048141 | 71.4286 | 60.1824 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.0914 | 58.1121 | oreochromis_niloticus | ENSONIP00000032766 | 64.2667 | 52.5333 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 56.6372 | 46.3127 | oreochromis_niloticus | ENSONIP00000028816 | 73.8462 | 60.3846 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 70.7965 | 57.8171 | oreochromis_niloticus | ENSONIP00000014128 | 73.1707 | 59.7561 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.0914 | 57.8171 | haplochromis_burtoni | ENSHBUP00000004955 | 73.4756 | 59.7561 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.3215 | 56.9322 | astatotilapia_calliptera | ENSACLP00000012188 | 76.7974 | 63.0719 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 70.5015 | 57.8171 | astatotilapia_calliptera | ENSACLP00000030630 | 76.8489 | 63.0225 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 57.2271 | 46.0177 | astatotilapia_calliptera | ENSACLP00000033980 | 69.7842 | 56.1151 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.6165 | 57.8171 | sparus_aurata | ENSSAUP00010057396 | 71.7325 | 59.5745 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 70.2065 | 58.1121 | lates_calcarifer | ENSLCAP00010025366 | 72.561 | 60.061 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 76.9911 | 64.0118 | xenopus_tropicalis | ENSXETP00000107486 | 81.0559 | 67.3913 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 81.1209 | 72.2714 | chrysemys_picta_bellii | ENSCPBP00000028854 | 85.6698 | 76.324 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 79.056 | 70.7965 | sphenodon_punctatus | ENSSPUP00000014127 | 84.0125 | 75.2351 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 79.351 | 70.2065 | crocodylus_porosus | ENSCPRP00005014769 | 84.0625 | 74.375 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 23.3038 | 17.6991 | laticauda_laticaudata | ENSLLTP00000018101 | 61.7188 | 46.875 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 74.6313 | 64.0118 | notechis_scutatus | ENSNSUP00000019069 | 80.3175 | 68.8889 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 77.5811 | 67.8466 | pseudonaja_textilis | ENSPTXP00000017914 | 82.1875 | 71.875 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 80.531 | 71.9764 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000002887 | 83.2317 | 74.3902 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 81.7109 | 71.9764 | gallus_gallus | ENSGALP00010012926 | 79.1429 | 69.7143 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 46.9027 | 39.823 | meleagris_gallopavo | ENSMGAP00000006664 | 58.0292 | 49.2701 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 75.5162 | 66.3717 | serinus_canaria | ENSSCAP00000019191 | 80.5031 | 70.7547 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 65.7817 | 60.767 | parus_major | ENSPMJP00000002949 | 94.4915 | 87.2881 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 60.177 | 54.2773 | anolis_carolinensis | ENSACAP00000024888 | 77.2727 | 69.697 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 21.5339 | 16.5192 | anolis_carolinensis | ENSACAP00000041464 | 59.3496 | 45.5285 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 55.1622 | 43.0678 | neolamprologus_brichardi | ENSNBRP00000008621 | 67.509 | 52.7076 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.0914 | 57.5221 | neolamprologus_brichardi | ENSNBRP00000003219 | 63.9257 | 51.7241 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 24.1888 | 21.5339 | naja_naja | ENSNNAP00000000802 | 85.4167 | 76.0417 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 22.7139 | 17.4041 | naja_naja | ENSNNAP00000000823 | 60.6299 | 46.4567 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 79.351 | 69.3215 | podarcis_muralis | ENSPMRP00000025264 | 78.4257 | 68.5131 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 67.5516 | 60.472 | geospiza_fortis | ENSGFOP00000004576 | 87.7395 | 78.5441 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 82.0059 | 73.1563 | taeniopygia_guttata | ENSTGUP00000024966 | 84.4985 | 75.3799 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.3864 | 58.9971 | erpetoichthys_calabaricus | ENSECRP00000021772 | 71.5976 | 59.1716 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 70.5015 | 58.4071 | kryptolebias_marmoratus | ENSKMAP00000015971 | 73.3129 | 60.7362 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 79.941 | 69.3215 | salvator_merianae | ENSSMRP00000024934 | 80.6548 | 69.9405 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.3864 | 59.292 | oryzias_javanicus | ENSOJAP00000008352 | 74.9226 | 62.2291 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 67.2566 | 54.5723 | amphilophus_citrinellus | ENSACIP00000013420 | 69.7248 | 56.5749 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.3215 | 58.7021 | amphiprion_percula | ENSAPEP00000018783 | 73.2087 | 61.9938 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 54.2773 | 44.2478 | oryzias_sinensis | ENSOSIP00000002523 | 85.9813 | 70.0935 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 63.7168 | 58.7021 | pelodiscus_sinensis | ENSPSIP00000013400 | 93.913 | 86.5217 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 81.1209 | 72.2714 | gopherus_evgoodei | ENSGEVP00005005269 | 85.6698 | 76.324 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 28.0236 | 22.1239 | chelonoidis_abingdonii | ENSCABP00000017566 | 67.8571 | 53.5714 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 62.8319 | 53.0973 | chelonoidis_abingdonii | ENSCABP00000004175 | 61.7391 | 52.1739 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 70.2065 | 58.9971 | seriola_dumerili | ENSSDUP00000025175 | 72.3404 | 60.7903 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.0266 | 57.2271 | cottoperca_gobio | ENSCGOP00000043990 | 71.3415 | 59.1463 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 65.4867 | 61.3569 | struthio_camelus_australis | ENSSCUP00000023409 | 94.4681 | 88.5106 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 81.1209 | 72.5664 | anser_brachyrhynchus | ENSABRP00000001165 | 82.8313 | 74.0964 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 67.5516 | 56.9322 | gasterosteus_aculeatus | ENSGACP00000024665 | 77.1044 | 64.9832 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 61.9469 | 50.7375 | cynoglossus_semilaevis | ENSCSEP00000015296 | 68.6274 | 56.2092 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 66.9617 | 56.3422 | poecilia_formosa | ENSPFOP00000000514 | 78.0069 | 65.6357 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 70.7965 | 59.587 | myripristis_murdjan | ENSMMDP00005025165 | 73.3945 | 61.7737 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 70.5015 | 58.4071 | sander_lucioperca | ENSSLUP00000043984 | 71.988 | 59.6386 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 80.531 | 72.5664 | strigops_habroptila | ENSSHBP00005022369 | 84.2593 | 75.9259 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.9115 | 58.7021 | amphiprion_ocellaris | ENSAOCP00000031507 | 73.8318 | 61.9938 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 81.1209 | 72.8614 | terrapene_carolina_triunguis | ENSTMTP00000022648 | 85.6698 | 76.947 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.3864 | 57.2271 | hucho_hucho | ENSHHUP00000003796 | 74.2331 | 59.5092 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.6165 | 56.9322 | betta_splendens | ENSBSLP00000004568 | 72.1713 | 59.0214 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.9115 | 57.5221 | betta_splendens | ENSBSLP00000006201 | 72.2561 | 59.4512 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.9764 | 59.882 | pygocentrus_nattereri | ENSPNAP00000014182 | 77.9553 | 64.8562 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 68.7316 | 56.3422 | anabas_testudineus | ENSATEP00000015224 | 71.4724 | 58.589 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 62.5369 | 48.0826 | ciona_savignyi | ENSCSAVP00000015534 | 70.1987 | 53.9735 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.0266 | 58.1121 | seriola_lalandi_dorsalis | ENSSLDP00000022048 | 73.125 | 61.5625 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 68.7316 | 56.0472 | labrus_bergylta | ENSLBEP00000032435 | 73.2704 | 59.7484 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 68.7316 | 56.0472 | labrus_bergylta | ENSLBEP00000020457 | 73.2704 | 59.7484 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 63.1268 | 49.5575 | pundamilia_nyererei | ENSPNYP00000005061 | 70.6271 | 55.4455 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.9115 | 57.2271 | mastacembelus_armatus | ENSMAMP00000033857 | 72.0365 | 58.9666 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.0914 | 58.7021 | stegastes_partitus | ENSSPAP00000009460 | 75.0779 | 61.9938 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.0914 | 57.5221 | oncorhynchus_tshawytscha | ENSOTSP00005088616 | 73.9264 | 59.816 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.0914 | 59.587 | acanthochromis_polyacanthus | ENSAPOP00000003286 | 75.0779 | 62.9283 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 75.5162 | 66.9617 | aquila_chrysaetos_chrysaetos | ENSACCP00020016084 | 77.5758 | 68.7879 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.9115 | 57.8171 | nothobranchius_furzeri | ENSNFUP00015007460 | 65.651 | 54.2936 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 46.6077 | 38.3481 | cyprinodon_variegatus | ENSCVAP00000012076 | 74.8815 | 61.6114 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.0914 | 58.9971 | denticeps_clupeoides | ENSDCDP00000006852 | 80.3333 | 66.6667 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 70.5015 | 57.2271 | larimichthys_crocea | ENSLCRP00005061623 | 66.205 | 53.7396 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 65.7817 | 55.7522 | takifugu_rubripes | ENSTRUP00000024191 | 72.8758 | 61.7647 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 41.5929 | 35.3982 | ficedula_albicollis | ENSFALP00000022492 | 67.7885 | 57.6923 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 31.2684 | 27.4336 | ficedula_albicollis | ENSFALP00000029276 | 89.0756 | 78.1513 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 70.7965 | 58.9971 | hippocampus_comes | ENSHCOP00000024626 | 73.3945 | 61.1621 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 65.7817 | 51.0324 | cyprinus_carpio_carpio | ENSCCRP00000101585 | 54.9261 | 42.6108 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 23.8938 | 20.649 | cyprinus_carpio_carpio | ENSCCRP00000118936 | 84.375 | 72.9167 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 76.6962 | 66.9617 | coturnix_japonica | ENSCJPP00005011102 | 71.8232 | 62.7072 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 60.767 | 49.5575 | poecilia_latipinna | ENSPLAP00000020892 | 67.3203 | 54.902 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.9115 | 56.0472 | sinocyclocheilus_grahami | ENSSGRP00000040980 | 76.4516 | 61.2903 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 70.5015 | 58.1121 | maylandia_zebra | ENSMZEP00005038062 | 76.8489 | 63.3441 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 61.0619 | 48.9676 | maylandia_zebra | ENSMZEP00005014626 | 69.2308 | 55.5184 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 63.1268 | 51.9174 | maylandia_zebra | ENSMZEP00005003466 | 64.6526 | 53.1722 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.3215 | 56.9322 | maylandia_zebra | ENSMZEP00005030008 | 76.7974 | 63.0719 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.0266 | 56.6372 | tetraodon_nigroviridis | ENSTNIP00000009904 | 76.7213 | 62.9508 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 72.2714 | 59.882 | paramormyrops_kingsleyae | ENSPKIP00000003430 | 71.2209 | 59.0116 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.0914 | 59.587 | scleropages_formosus | ENSSFOP00015011752 | 73.7003 | 61.7737 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 74.6313 | 63.4218 | leptobrachium_leishanense | ENSLLEP00000000207 | 78.0864 | 66.358 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 69.0266 | 56.9322 | scophthalmus_maximus | ENSSMAP00000011804 | 78.2609 | 64.5485 |
| ENSG00000148090 | homo_sapiens | ENSP00000364883 | 71.6814 | 59.587 | oryzias_melastigma | ENSOMEP00000005867 | 75.2322 | 62.5387 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0003730 | enables mRNA 3-UTR binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0004300 | enables enoyl-CoA hydratase activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0004490 | enables methylglutaconyl-CoA hydratase activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005739 | is_active_in mitochondrion | 2 | IBA,ISS | Homo_sapiens(9606) | Component |
| GO:0005759 | located_in mitochondrial matrix | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0006552 | involved_in leucine catabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006635 | involved_in fatty acid beta-oxidation | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0050011 | enables itaconyl-CoA hydratase activity | 1 | IEA | Homo_sapiens(9606) | Function |