RBP Type
Non-canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
TF
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000147596 Gene ID
63978 Accession
14001
Symbol
PRDM14
Alias
PFM11
Full Name
PR/SET domain 14
Status
Confidence
Length
20043 bases
Strand
Minus strand
Position
8 : 70051651 - 70071693
RNA binding domain
N.A.
Summary
This gene encodes a member of the PRDI-BF1 and RIZ homology domain containing (PRDM) family of transcriptional regulators. The encoded protein may possess histone methyltransferase activity and plays a critical role in cell pluripotency by suppressing the expression of differentiation marker genes. Expression of this gene may play a role in breast cancer. [provided by RefSeq, Dec 2011]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
28423353 Silencing PRDM14 expression by an innovative RNAi therapy inhibits stemness, tumorigenicity, and metastasis of breast cancer.
Hiroaki Taniguchi
2017-07-18
Oncotarget
Name
Transcript ID
bp
Protein
Translation ID
PRDM14-201
ENST00000276594
2269
571aa
ENSP00000276594
PRDM14-202
ENST00000426346
422
99aa
ENSP00000408653
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
76695 Testicular Neoplasms 5E-8 23666240
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
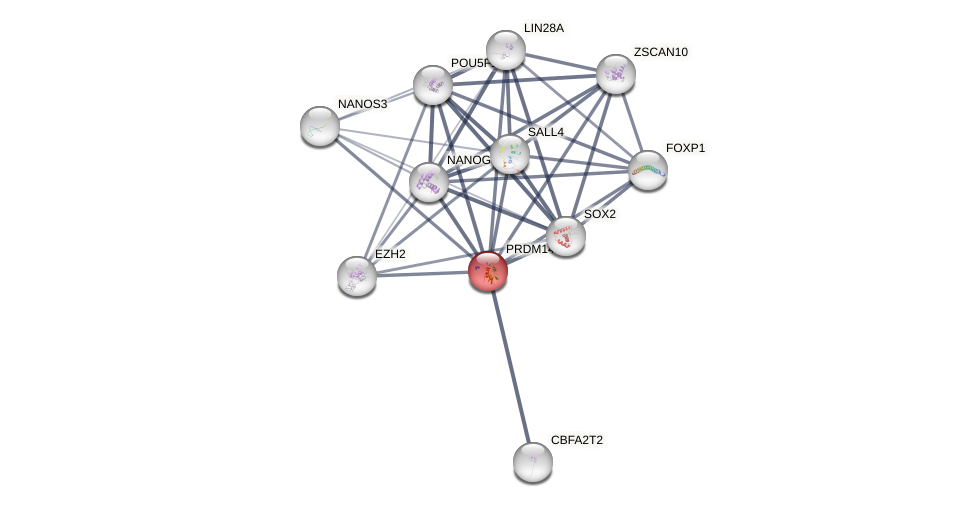
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000122 involved_in negative regulation of transcription by RNA polymerase II 1 IEA Homo_sapiens(9606) Process GO:0000902 involved_in cell morphogenesis 1 IEA Homo_sapiens(9606) Process GO:0000977 enables RNA polymerase II transcription regulatory region sequence-specific DNA binding 1 IBA Homo_sapiens(9606) Function GO:0001708 involved_in cell fate specification 1 IEA Homo_sapiens(9606) Process GO:0001827 involved_in inner cell mass cell fate commitment 1 IEA Homo_sapiens(9606) Process GO:0003723 enables RNA binding 1 IEA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 is_active_in nucleus 1 IBA Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 2 IDA,TAS Homo_sapiens(9606) Component GO:0006357 involved_in regulation of transcription by RNA polymerase II 1 IBA Homo_sapiens(9606) Process GO:0007281 involved_in germ cell development 1 IEA Homo_sapiens(9606) Process GO:0007566 involved_in embryo implantation 1 IEA Homo_sapiens(9606) Process GO:0008168 enables methyltransferase activity 1 IEA Homo_sapiens(9606) Function GO:0008543 involved_in fibroblast growth factor receptor signaling pathway 1 IEA Homo_sapiens(9606) Process GO:0009566 involved_in fertilization 1 IEA Homo_sapiens(9606) Process GO:0030718 involved_in germ-line stem cell population maintenance 1 IEA Homo_sapiens(9606) Process GO:0031490 enables chromatin DNA binding 1 IEA Homo_sapiens(9606) Function GO:0034972 involved_in histone H3-R26 methylation 1 IBA Homo_sapiens(9606) Process GO:0040037 involved_in negative regulation of fibroblast growth factor receptor signaling pathway 1 IEA Homo_sapiens(9606) Process GO:0044030 involved_in regulation of DNA methylation 1 IEA Homo_sapiens(9606) Process GO:0046872 enables metal ion binding 1 IEA Homo_sapiens(9606) Function GO:0048873 involved_in homeostasis of number of cells within a tissue 1 IEA Homo_sapiens(9606) Process GO:0060817 involved_in inactivation of paternal X chromosome 1 IEA Homo_sapiens(9606) Process GO:1902093 involved_in positive regulation of flagellated sperm motility 1 IEA Homo_sapiens(9606) Process GO:1902459 involved_in positive regulation of stem cell population maintenance 1 IEA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.