Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000143799 | Gene ID | 142 | Accession | 270 |
| Symbol | PARP1 | Alias | PARP;PARS;PPOL;ADPRT;ARTD1;ADPRT1;PARP-1;ADPRT1;pADPRT-1;Poly-PARP | Full Name | poly(ADP-ribose) polymerase 1 |
| Status | Confidence | Length | 47410 bases | Strand | Minus strand |
| Position | 1 : 226360691 - 226408100 | RNA binding domain | PARP | ||
| Summary | This gene encodes a chromatin-associated enzyme, poly(ADP-ribosyl)transferase, which modifies various nuclear proteins by poly(ADP-ribosyl)ation. The modification is dependent on DNA and is involved in the regulation of various important cellular processes such as differentiation, proliferation, and tumor transformation and also in the regulation of the molecular events involved in the recovery of cell from DNA damage. In addition, this enzyme may be the site of mutation in Fanconi anemia, and may participate in the pathophysiology of type I diabetes. [provided by RefSeq, Jul 2008] | ||||
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| PARP1-215 | ENST00000677189 | 2348 | No protein | - |
| PARP1-220 | ENST00000677985 | 2285 | No protein | - |
| PARP1-223 | ENST00000678288 | 2420 | No protein | - |
| PARP1-222 | ENST00000678226 | 3274 | No protein | - |
| PARP1-210 | ENST00000676481 | 1371 | No protein | - |
| PARP1-203 | ENST00000366794 | 3978 | 1014aa | ENSP00000355759 |
| PARP1-224 | ENST00000678560 | 4255 | 100aa | ENSP00000503293 |
| PARP1-219 | ENST00000677884 | 4620 | No protein | - |
| PARP1-216 | ENST00000677203 | 3850 | 971aa | ENSP00000503396 |
| PARP1-211 | ENST00000676565 | 3577 | No protein | - |
| PARP1-221 | ENST00000678144 | 3719 | 308aa | ENSP00000504430 |
| PARP1-214 | ENST00000677091 | 3768 | 369aa | ENSP00000504745 |
| PARP1-225 | ENST00000678781 | 5430 | No protein | - |
| PARP1-212 | ENST00000676685 | 6210 | No protein | - |
| PARP1-226 | ENST00000679276 | 4918 | No protein | - |
| PARP1-209 | ENST00000498787 | 5629 | No protein | - |
| PARP1-213 | ENST00000676709 | 4764 | No protein | - |
| PARP1-207 | ENST00000490921 | 3165 | No protein | - |
| PARP1-204 | ENST00000463968 | 830 | No protein | - |
| PARP1-208 | ENST00000491816 | 416 | No protein | - |
| PARP1-205 | ENST00000468608 | 438 | No protein | - |
| PARP1-217 | ENST00000677374 | 5697 | No protein | - |
| PARP1-218 | ENST00000677815 | 3770 | No protein | - |
| PARP1-206 | ENST00000469663 | 542 | No protein | - |
| PARP1-202 | ENST00000366792 | 553 | No protein | - |
| PARP1-201 | ENST00000366790 | 3358 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 75382 | Melanoma | 9E-8 | 21983785 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
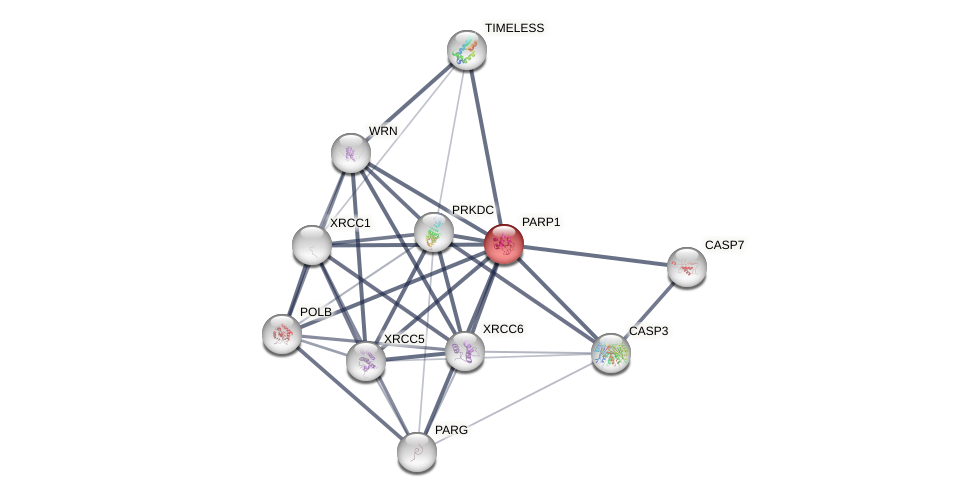
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000143799 | homo_sapiens | ENSP00000392972 | 60 | 39.1228 | homo_sapiens | ENSP00000355759 | 33.7278 | 21.9921 |
| ENSG00000143799 | homo_sapiens | ENSP00000381740 | 49.1557 | 32.0826 | homo_sapiens | ENSP00000355759 | 25.8383 | 16.8639 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 92.8994 | 92.2091 | nomascus_leucogenys | ENSNLEP00000016881 | 99.3671 | 98.6287 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 99.8028 | 99.4083 | pan_paniscus | ENSPPAP00000036168 | 99.8028 | 99.4083 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 99.8028 | 99.4083 | pongo_abelii | ENSPPYP00000000162 | 99.8028 | 99.4083 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 99.8028 | 99.4083 | pan_troglodytes | ENSPTRP00000003457 | 99.8028 | 99.4083 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 100 | 99.6055 | gorilla_gorilla | ENSGGOP00000025582 | 100 | 99.6055 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 89.645 | 84.0237 | ornithorhynchus_anatinus | ENSOANP00000002367 | 81.9657 | 76.826 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 89.7436 | 83.1361 | sarcophilus_harrisii | ENSSHAP00000002240 | 91 | 84.3 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 61.4398 | 57.8895 | notamacropus_eugenii | ENSMEUP00000007053 | 80.6995 | 76.0363 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 55.4241 | 53.5503 | choloepus_hoffmanni | ENSCHOP00000003145 | 55.5336 | 53.6561 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 91.8146 | 88.3629 | dasypus_novemcinctus | ENSDNOP00000009366 | 95.9794 | 92.3711 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 92.8994 | 88.9546 | erinaceus_europaeus | ENSEEUP00000011219 | 92.7165 | 88.7795 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 73.7673 | 69.428 | echinops_telfairi | ENSETEP00000010750 | 73.2615 | 68.952 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 96.6469 | 95.069 | callithrix_jacchus | ENSCJAP00000072405 | 94.2308 | 92.6923 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 99.211 | 98.2249 | cercocebus_atys | ENSCATP00000031757 | 99.211 | 98.2249 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 99.0138 | 98.1262 | macaca_fascicularis | ENSMFAP00000005740 | 99.0138 | 98.1262 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 99.0138 | 98.1262 | macaca_mulatta | ENSMMUP00000028737 | 99.0138 | 98.1262 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 99.211 | 98.2249 | macaca_nemestrina | ENSMNEP00000020544 | 99.211 | 98.2249 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 99.1124 | 98.1262 | papio_anubis | ENSPANP00000003629 | 94.722 | 93.7795 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.3373 | 96.2525 | mandrillus_leucophaeus | ENSMLEP00000002973 | 97.0501 | 95.9685 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 91.8146 | 88.5602 | tursiops_truncatus | ENSTTRP00000000174 | 92.087 | 88.8229 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 92.8994 | 90.3353 | vulpes_vulpes | ENSVVUP00000002030 | 96.9136 | 94.2387 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 95.858 | 92.5049 | mus_spretus | MGP_SPRETEiJ_P0022674 | 95.858 | 92.5049 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.2387 | 95.1677 | equus_asinus | ENSEASP00005047389 | 97.2387 | 95.1677 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 93.4911 | 89.1519 | capra_hircus | ENSCHIP00000019007 | 92.9412 | 88.6274 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 91.8146 | 87.7712 | loxodonta_africana | ENSLAFP00000006575 | 95.5852 | 91.3758 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 95.858 | 92.4063 | panthera_leo | ENSPLOP00000008694 | 95.9526 | 92.4975 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 88.6588 | 85.6016 | ailuropoda_melanoleuca | ENSAMEP00000015307 | 96.2527 | 92.9336 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 94.7732 | 90.8284 | bos_indicus_hybrid | ENSBIXP00005043645 | 94.5866 | 90.6496 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.6331 | 94.5759 | oryctolagus_cuniculus | ENSOCUP00000015305 | 97.7295 | 94.6693 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.2387 | 95.1677 | equus_caballus | ENSECAP00000031918 | 90.7919 | 88.8582 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.14 | 94.3787 | canis_lupus_familiaris | ENSCAFP00845011348 | 97.14 | 94.3787 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 92.2091 | 88.7574 | panthera_pardus | ENSPPRP00000008865 | 96.0946 | 92.4974 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.929 | 95.2663 | marmota_marmota_marmota | ENSMMMP00000027565 | 97.929 | 95.2663 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.14 | 94.0828 | balaenoptera_musculus | ENSBMSP00010003003 | 97.2359 | 94.1757 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 96.4497 | 92.998 | cavia_porcellus | ENSCPOP00000014109 | 96.7359 | 93.274 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 92.6036 | 89.4477 | felis_catus | ENSFCAP00000005366 | 95.9142 | 92.6456 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 92.998 | 90.0394 | ursus_americanus | ENSUAMP00000018127 | 96.7179 | 93.641 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 91.3215 | 84.8126 | monodelphis_domestica | ENSMODP00000007535 | 91.4117 | 84.8963 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 94.7732 | 90.6312 | bison_bison_bison | ENSBBBP00000019055 | 94.8667 | 90.7206 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 96.9428 | 93.6884 | monodon_monoceros | ENSMMNP00015011712 | 97.0385 | 93.7808 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.0414 | 94.5759 | sus_scrofa | ENSSSCP00000022104 | 96.7552 | 94.297 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 93.9842 | 92.1105 | microcebus_murinus | ENSMICP00000015556 | 97.4438 | 95.501 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 94.6746 | 90.4339 | octodon_degus | ENSODEP00000009928 | 95.1437 | 90.8821 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 95.4635 | 91.3215 | jaculus_jaculus | ENSJJAP00000023860 | 95.4635 | 91.3215 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 95.1677 | 91.4201 | ovis_aries_rambouillet | ENSOARP00020024506 | 94.9803 | 91.2402 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.3195 | 80.3748 | ursus_maritimus | ENSUMAP00000020551 | 88.785 | 84.6314 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.2387 | 93.5897 | ochotona_princeps | ENSOPRP00000010706 | 97.2387 | 93.5897 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.0414 | 93.787 | delphinapterus_leucas | ENSDLEP00000002220 | 97.1372 | 93.8796 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.2387 | 94.5759 | physeter_catodon | ENSPCTP00005001476 | 97.3346 | 94.6693 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 95.7594 | 92.5049 | mus_caroli | MGP_CAROLIEiJ_P0021609 | 95.7594 | 92.5049 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 95.858 | 92.5049 | mus_musculus | ENSMUSP00000027777 | 95.858 | 92.5049 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 92.8008 | 87.7712 | dipodomys_ordii | ENSDORP00000004897 | 94.1 | 89 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 96.5483 | 92.8008 | mustela_putorius_furo | ENSMPUP00000004550 | 96.6436 | 92.8924 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 96.5483 | 94.8718 | aotus_nancymaae | ENSANAP00000029984 | 94.1346 | 92.5 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 96.3511 | 94.3787 | saimiri_boliviensis_boliviensis | ENSSBOP00000001954 | 91.9962 | 90.113 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 80.8679 | 78.7968 | vicugna_pacos | ENSVPAP00000010630 | 81.1078 | 79.0307 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 69.1321 | 68.4418 | chlorocebus_sabaeus | ENSCSAP00000012419 | 98.8717 | 97.8843 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 77.712 | 75.9369 | tupaia_belangeri | ENSTBEP00000002064 | 87.4584 | 85.4606 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 95.858 | 93.0966 | mesocricetus_auratus | ENSMAUP00000006331 | 95.9526 | 93.1885 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.14 | 93.5897 | phocoena_sinus | ENSPSNP00000004870 | 97.2359 | 93.6821 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 95.2663 | 92.7022 | camelus_dromedarius | ENSCDRP00005005781 | 94.8919 | 92.3379 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 88.856 | 86.6864 | carlito_syrichta | ENSTSYP00000003407 | 98.0414 | 95.6474 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 77.8107 | 73.8659 | panthera_tigris_altaica | ENSPTIP00000007494 | 89.9658 | 85.4048 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 95.3649 | 91.8146 | rattus_norvegicus | ENSRNOP00000004232 | 95.3649 | 91.8146 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.4359 | 94.8718 | prolemur_simus | ENSPSMP00000006208 | 97.6285 | 95.0593 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 95.7594 | 92.7022 | microtus_ochrogaster | ENSMOCP00000014984 | 95.8539 | 92.7937 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 52.4655 | 50.9862 | sorex_araneus | ENSSARP00000008541 | 57.9521 | 56.3181 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 94.8718 | 92.0118 | peromyscus_maniculatus_bairdii | ENSPEMP00000023071 | 95.8167 | 92.9283 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 10.6509 | 9.76331 | bos_mutus | ENSBMUP00000011748 | 78.8321 | 72.2628 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 67.6529 | 63.5108 | bos_mutus | ENSBMUP00000020098 | 84.6914 | 79.5062 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 94.0828 | 90.7298 | mus_spicilegus | ENSMSIP00000008396 | 95.7831 | 92.3695 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 96.0552 | 93.1953 | cricetulus_griseus_chok1gshd | ENSCGRP00001021836 | 96.15 | 93.2873 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 78.8955 | 75.1479 | pteropus_vampyrus | ENSPVAP00000005249 | 79.1296 | 75.3709 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 74.359 | 68.146 | myotis_lucifugus | ENSMLUP00000011191 | 83.4994 | 76.5227 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 91.1243 | 84.9112 | vombatus_ursinus | ENSVURP00010021103 | 91.3043 | 85.079 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 91.1243 | 84.9112 | vombatus_ursinus | ENSVURP00010011507 | 91.3043 | 85.079 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 96.8442 | 93.3925 | rhinolophus_ferrumequinum | ENSRFEP00010021137 | 96.8442 | 93.3925 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 96.5483 | 92.7022 | neovison_vison | ENSNVIP00000018942 | 96.6436 | 92.7937 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 93.3925 | 89.0533 | bos_taurus | ENSBTAP00000059409 | 92.2103 | 87.926 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 95.6608 | 91.716 | heterocephalus_glaber_female | ENSHGLP00000005446 | 96.2302 | 92.2619 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 98.9152 | 97.929 | rhinopithecus_bieti | ENSRBIP00000043459 | 98.9152 | 97.929 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.14 | 94.2801 | canis_lupus_dingo | ENSCAFP00020003677 | 97.14 | 94.2801 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.8304 | 95.4635 | sciurus_vulgaris | ENSSVLP00005012835 | 97.9269 | 95.5577 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 91.3215 | 84.714 | phascolarctos_cinereus | ENSPCIP00000007697 | 91.3215 | 84.714 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 56.213 | 53.1558 | procavia_capensis | ENSPCAP00000005300 | 56.2685 | 53.2083 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 96.0552 | 92.6036 | nannospalax_galili | ENSNGAP00000003404 | 96.15 | 92.695 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 94.7732 | 90.927 | bos_grunniens | ENSBGRP00000008626 | 94.5866 | 90.748 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 96.1538 | 93.0966 | chinchilla_lanigera | ENSCLAP00000009568 | 96.2488 | 93.1885 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 95.069 | 91.5187 | cervus_hanglu_yarkandensis | ENSCHYP00000014284 | 94.8819 | 91.3386 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 96.8442 | 93.5897 | catagonus_wagneri | ENSCWAP00000003788 | 96.4637 | 93.222 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 98.0276 | 95.5621 | ictidomys_tridecemlineatus | ENSSTOP00000014322 | 98.0276 | 95.5621 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 96.9428 | 93.4911 | otolemur_garnettii | ENSOGAP00000016342 | 96.6568 | 93.2153 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 98.4221 | 96.9428 | cebus_imitator | ENSCCAP00000030861 | 98.5192 | 97.0385 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 88.2643 | 86.0947 | urocitellus_parryii | ENSUPAP00010002753 | 97.6009 | 95.2017 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 95.2663 | 91.716 | moschus_moschiferus | ENSMMSP00000025353 | 95.0787 | 91.5354 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 98.9152 | 97.929 | rhinopithecus_roxellana | ENSRROP00000009714 | 98.9152 | 97.929 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 9.46746 | 9.07298 | mus_pahari | MGP_PahariEiJ_P0076859 | 97.9592 | 93.8775 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 97.6331 | 94.7732 | propithecus_coquereli | ENSPCOP00000010883 | 97.6331 | 94.7732 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 50.0986 | 32.643 | caenorhabditis_elegans | Y71F9AL.18a.2 | 53.7566 | 35.0265 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 59.3688 | 41.4201 | drosophila_melanogaster | FBpp0112608 | 60.5634 | 42.2535 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 36.2919 | 27.9093 | ciona_intestinalis | ENSCINP00000034447 | 77.9661 | 59.9576 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 21.7949 | 16.7653 | petromyzon_marinus | ENSPMAP00000010510 | 63.8728 | 49.1329 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 73.2742 | 58.4813 | eptatretus_burgeri | ENSEBUP00000008996 | 71.58 | 57.1291 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 80.0789 | 67.0611 | callorhinchus_milii | ENSCMIP00000013962 | 78.4541 | 65.7005 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 70.4142 | 59.7633 | latimeria_chalumnae | ENSLACP00000003056 | 70.6231 | 59.9407 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.925 | 73.0769 | lepisosteus_oculatus | ENSLOCP00000019452 | 84.3409 | 73.439 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.5306 | 71.0059 | clupea_harengus | ENSCHAP00000017150 | 82.7148 | 70.3125 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.1223 | 71.499 | danio_rerio | ENSDARP00000008364 | 84.2053 | 71.5696 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 81.854 | 68.9349 | carassius_auratus | ENSCARP00000102010 | 78.7476 | 66.3188 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.6154 | 71.0059 | carassius_auratus | ENSCARP00000108940 | 84.3658 | 70.7965 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 81.3609 | 68.5404 | carassius_auratus | ENSCARP00000087834 | 80.0194 | 67.4103 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.2347 | 70.217 | astyanax_mexicanus | ENSAMXP00000006642 | 82.6641 | 69.7355 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.3333 | 69.7239 | ictalurus_punctatus | ENSIPUP00000030007 | 83.0059 | 69.4499 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.432 | 71.7949 | esox_lucius | ENSELUP00000017644 | 83.6795 | 72.0079 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.1361 | 70.8087 | electrophorus_electricus | ENSEEEP00000030073 | 83.3004 | 70.9486 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.6292 | 70.5128 | oncorhynchus_kisutch | ENSOKIP00005047921 | 83.4646 | 70.374 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.1361 | 70.1183 | oncorhynchus_mykiss | ENSOMYP00000053004 | 83.2182 | 70.1876 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 82.7416 | 70.1183 | salmo_salar | ENSSSAP00000053320 | 83.0693 | 70.396 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 81.1637 | 68.2446 | salmo_trutta | ENSSTUP00000062523 | 81.0837 | 68.1773 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 82.9389 | 70.1183 | gadus_morhua | ENSGMOP00000028410 | 83.0207 | 70.1876 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.3195 | 70.7101 | fundulus_heteroclitus | ENSFHEP00000000402 | 84.2365 | 70.6404 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.1223 | 71.3018 | poecilia_reticulata | ENSPREP00000001596 | 84.0394 | 71.2315 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.1223 | 70.3156 | xiphophorus_maculatus | ENSXMAP00000003639 | 84.0394 | 70.2463 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.8264 | 70.4142 | oryzias_latipes | ENSORLP00000017176 | 83.7438 | 70.3448 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.7278 | 70.0197 | cyclopterus_lumpus | ENSCLMP00005021923 | 83.7278 | 70.0197 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.3195 | 71.8935 | oreochromis_niloticus | ENSONIP00000074983 | 83.5777 | 71.261 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.8126 | 72.0907 | haplochromis_burtoni | ENSHBUP00000008939 | 84.8126 | 72.0907 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.1223 | 71.6963 | astatotilapia_calliptera | ENSACLP00000022919 | 85.3854 | 72.7728 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.1223 | 71.9921 | sparus_aurata | ENSSAUP00010049439 | 84.1223 | 71.9921 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.4181 | 71.4004 | lates_calcarifer | ENSLCAP00010021233 | 84.4181 | 71.4004 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 86.0947 | 74.9507 | xenopus_tropicalis | ENSXETP00000081175 | 86.1797 | 75.0247 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 88.5602 | 80.3748 | chrysemys_picta_bellii | ENSCPBP00000016211 | 88.8229 | 80.6133 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.6154 | 76.43 | sphenodon_punctatus | ENSSPUP00000010080 | 87.1951 | 78.7602 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 87.7712 | 78.9941 | crocodylus_porosus | ENSCPRP00005004756 | 88.0317 | 79.2285 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 70.0197 | 61.144 | laticauda_laticaudata | ENSLLTP00000014515 | 88.4184 | 77.2105 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 86.1933 | 75.1479 | notechis_scutatus | ENSNSUP00000013158 | 86.6204 | 75.5203 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 86.2919 | 75.1479 | pseudonaja_textilis | ENSPTXP00000018539 | 86.6337 | 75.4455 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 85.4043 | 76.6272 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000014006 | 87.2984 | 78.3266 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 87.7712 | 77.712 | gallus_gallus | ENSGALP00010024778 | 85.3308 | 75.5513 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.9112 | 76.0355 | meleagris_gallopavo | ENSMGAP00000019530 | 88.8545 | 79.5666 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 86.5878 | 76.7258 | serinus_canaria | ENSSCAP00000021487 | 87.2763 | 77.336 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 88.2643 | 79.1913 | parus_major | ENSPMJP00000015554 | 88.2643 | 79.1913 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 87.1795 | 77.3176 | anolis_carolinensis | ENSACAP00000036234 | 87.8728 | 77.9324 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.2209 | 71.5976 | neolamprologus_brichardi | ENSNBRP00000022802 | 85.4855 | 72.6727 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 86.4892 | 75.9369 | naja_naja | ENSNNAP00000000074 | 86.7458 | 76.1622 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 86.9822 | 77.8107 | podarcis_muralis | ENSPMRP00000003666 | 87.4133 | 78.1962 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.7278 | 74.1617 | geospiza_fortis | ENSGFOP00000010753 | 80.5503 | 71.3473 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.9112 | 76.0355 | taeniopygia_guttata | ENSTGUP00000004425 | 88.5802 | 79.321 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 7.79093 | 6.50888 | erpetoichthys_calabaricus | ENSECRP00000005851 | 82.2917 | 68.75 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.0237 | 71.6963 | kryptolebias_marmoratus | ENSKMAP00000015581 | 83.9409 | 71.6256 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 87.3767 | 77.6134 | salvator_merianae | ENSSMRP00000001376 | 87.2906 | 77.5369 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.8264 | 70.4142 | oryzias_javanicus | ENSOJAP00000034543 | 83.7438 | 70.3448 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.2347 | 70.217 | amphilophus_citrinellus | ENSACIP00000007255 | 83.8133 | 70.7051 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.8264 | 71.2032 | dicentrarchus_labrax | ENSDLAP00005042924 | 83.6614 | 71.063 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.4181 | 71.6963 | amphiprion_percula | ENSAPEP00000006782 | 84.4181 | 71.6963 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.925 | 70.3156 | oryzias_sinensis | ENSOSIP00000017376 | 83.8424 | 70.2463 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.8126 | 76.3314 | pelodiscus_sinensis | ENSPSIP00000018712 | 88.2051 | 79.3846 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 88.4615 | 79.5858 | gopherus_evgoodei | ENSGEVP00005020856 | 88.724 | 79.822 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 88.4615 | 79.783 | chelonoidis_abingdonii | ENSCABP00000003267 | 88.724 | 80.0198 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.0237 | 70.7101 | seriola_dumerili | ENSSDUP00000022913 | 84.0237 | 70.7101 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.1361 | 69.5266 | cottoperca_gobio | ENSCGOP00000040599 | 82.8909 | 69.3215 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 86.0947 | 76.6272 | struthio_camelus_australis | ENSSCUP00000004103 | 85.3372 | 75.9531 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 87.9684 | 79.2899 | anser_brachyrhynchus | ENSABRP00000019463 | 88.4044 | 79.6829 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.0237 | 69.8225 | gasterosteus_aculeatus | ENSGACP00000014960 | 83.8583 | 69.685 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.3195 | 70.4142 | cynoglossus_semilaevis | ENSCSEP00000032212 | 84.2365 | 70.3448 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.2347 | 70.6114 | poecilia_formosa | ENSPFOP00000002611 | 83.1527 | 70.5419 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.432 | 70.9073 | myripristis_murdjan | ENSMMDP00005045913 | 84.7695 | 72.0441 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.432 | 70.7101 | sander_lucioperca | ENSSLUP00000057024 | 83.2677 | 70.5709 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 88.3629 | 79.1913 | strigops_habroptila | ENSSHBP00005023755 | 88.6251 | 79.4263 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.4181 | 71.6963 | amphiprion_ocellaris | ENSAOCP00000030156 | 84.4181 | 71.6963 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 88.5602 | 80.4734 | terrapene_carolina_triunguis | ENSTMTP00000019783 | 88.8229 | 80.7122 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 10.4536 | 8.38264 | hucho_hucho | ENSHHUP00000007390 | 75.1773 | 60.2837 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 82.4458 | 70.6114 | betta_splendens | ENSBSLP00000041388 | 82.4458 | 70.6114 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.4181 | 71.4004 | pygocentrus_nattereri | ENSPNAP00000007761 | 83.8394 | 70.9109 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.2347 | 70.4142 | anabas_testudineus | ENSATEP00000053525 | 82.3415 | 69.6585 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 16.4694 | 13.1164 | ciona_savignyi | ENSCSAVP00000007789 | 78.7736 | 62.7358 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.925 | 70.7101 | seriola_lalandi_dorsalis | ENSSLDP00000008111 | 83.925 | 70.7101 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.925 | 71.3018 | labrus_bergylta | ENSLBEP00000017409 | 84.0079 | 71.3722 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.1223 | 71.6963 | pundamilia_nyererei | ENSPNYP00000012848 | 85.3854 | 72.7728 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.4181 | 71.6963 | mastacembelus_armatus | ENSMAMP00000030646 | 84.7525 | 71.9802 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 82.0513 | 69.5266 | stegastes_partitus | ENSSPAP00000025320 | 83.1169 | 70.4296 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 82.9389 | 69.8225 | oncorhynchus_tshawytscha | ENSOTSP00005047660 | 82.8571 | 69.7537 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 25.7396 | 21.499 | oncorhynchus_tshawytscha | ENSOTSP00005057628 | 80.805 | 67.4923 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.8264 | 71.0059 | acanthochromis_polyacanthus | ENSAPOP00000004749 | 83.8264 | 71.0059 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 88.2643 | 79.1913 | aquila_chrysaetos_chrysaetos | ENSACCP00020018942 | 88.2643 | 79.1913 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 82.643 | 69.6252 | nothobranchius_furzeri | ENSNFUP00015009672 | 83.1349 | 70.0397 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 80.572 | 67.6529 | cyprinodon_variegatus | ENSCVAP00000031778 | 83.4525 | 70.0715 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.6292 | 71.5976 | denticeps_clupeoides | ENSDCDP00000059409 | 83.3825 | 71.3864 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.0237 | 71.5976 | larimichthys_crocea | ENSLCRP00005014865 | 84.0237 | 71.5976 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 82.2485 | 69.0335 | takifugu_rubripes | ENSTRUP00000049295 | 79.6562 | 66.8577 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 85.7002 | 76.43 | ficedula_albicollis | ENSFALP00000021150 | 86.2959 | 76.9613 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 82.9389 | 70.3156 | hippocampus_comes | ENSHCOP00000023234 | 82.451 | 69.902 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.0237 | 70.6114 | cyprinus_carpio_carpio | ENSCCRP00000041512 | 69.7218 | 58.5925 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.0237 | 71.3018 | cyprinus_carpio_carpio | ENSCCRP00000073342 | 83.8583 | 71.1614 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 87.2781 | 77.3176 | coturnix_japonica | ENSCJPP00005006708 | 86.5103 | 76.6373 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.2209 | 71.1045 | poecilia_latipinna | ENSPLAP00000000146 | 84.1379 | 71.0345 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 71.8935 | 60.2564 | sinocyclocheilus_grahami | ENSSGRP00000078252 | 76.7368 | 64.3158 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 74.7534 | 62.5247 | sinocyclocheilus_grahami | ENSSGRP00000059473 | 83.4802 | 69.8238 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.8126 | 72.288 | maylandia_zebra | ENSMZEP00005022593 | 84.8126 | 72.288 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.3333 | 70.3156 | tetraodon_nigroviridis | ENSTNIP00000019481 | 82.762 | 69.8335 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 82.7416 | 71.1045 | paramormyrops_kingsleyae | ENSPKIP00000021872 | 82.9871 | 71.3155 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 83.7278 | 71.8935 | scleropages_formosus | ENSSFOP00015031268 | 83.6453 | 71.8227 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 85.2071 | 71.8935 | leptobrachium_leishanense | ENSLLEP00000024605 | 85.4599 | 72.1068 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 84.3195 | 71.2032 | scophthalmus_maximus | ENSSMAP00000062055 | 84.3195 | 71.2032 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 8.1854 | 7.00197 | oryzias_melastigma | ENSOMEP00000006733 | 79.8077 | 68.2692 |
| ENSG00000143799 | homo_sapiens | ENSP00000355759 | 32.9389 | 28.4024 | oryzias_melastigma | ENSOMEP00000006747 | 84.557 | 72.9114 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000122 | involved_in negative regulation of transcription by RNA polymerase II | 2 | IDA,TAS | Homo_sapiens(9606) | Process |
| GO:0000723 | involved_in telomere maintenance | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0000781 | located_in chromosome, telomeric region | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0000785 | part_of chromatin | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0003677 | enables DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003682 | enables chromatin binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003684 | enables damaged DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0003950 | enables NAD+ ADP-ribosyltransferase activity | 4 | IBA,IDA,IMP,TAS | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 2 | IDA | Homo_sapiens(9606) | Component |
| GO:0005635 | located_in nuclear envelope | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005667 | part_of transcription regulator complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005730 | is_active_in nucleolus | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005739 | located_in mitochondrion | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | is_active_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0006281 | involved_in DNA repair | 2 | IDA,TAS | Homo_sapiens(9606) | Process |
| GO:0006302 | involved_in double-strand break repair | 3 | IBA,IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0006366 | involved_in transcription by RNA polymerase II | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006915 | involved_in apoptotic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006974 | involved_in DNA damage response | 3 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0007005 | acts_upstream_of_or_within mitochondrion organization | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0007179 | involved_in transforming growth factor beta receptor signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008270 | enables zinc ion binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0010332 | involved_in response to gamma radiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010613 | involved_in positive regulation of cardiac muscle hypertrophy | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0016020 | located_in membrane | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0016051 | involved_in carbohydrate biosynthetic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0016540 | involved_in protein autoprocessing | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0016604 | located_in nuclear body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0016779 | enables nucleotidyltransferase activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0019899 | enables enzyme binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0019901 | enables protein kinase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0023019 | involved_in signal transduction involved in regulation of gene expression | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0030225 | involved_in macrophage differentiation | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0030331 | enables nuclear estrogen receptor binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0030592 | involved_in DNA ADP-ribosylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0031491 | enables nucleosome binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0031625 | enables ubiquitin protein ligase binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0032042 | acts_upstream_of_or_within mitochondrial DNA metabolic process | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032786 | involved_in positive regulation of DNA-templated transcription, elongation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032869 | involved_in cellular response to insulin stimulus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032880 | acts_upstream_of_or_within regulation of protein localization | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032991 | part_of protein-containing complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0032993 | part_of protein-DNA complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0033148 | involved_in positive regulation of intracellular estrogen receptor signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0034244 | involved_in negative regulation of transcription elongation by RNA polymerase II | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0034599 | acts_upstream_of_or_within cellular response to oxidative stress | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0034644 | involved_in cellular response to UV | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0035861 | is_active_in site of double-strand break | 2 | IDA | Homo_sapiens(9606) | Component |
| GO:0036211 | acts_upstream_of_or_within protein modification process | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0042802 | enables identical protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0042803 | enables protein homodimerization activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0042826 | enables histone deacetylase binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0043123 | involved_in positive regulation of I-kappaB kinase/NF-kappaB signaling | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0043504 | acts_upstream_of_or_within mitochondrial DNA repair | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0043596 | is_active_in nuclear replication fork | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0044030 | involved_in regulation of DNA methylation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045087 | involved_in innate immune response | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045188 | involved_in regulation of circadian sleep/wake cycle, non-REM sleep | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0045824 | involved_in negative regulation of innate immune response | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0045892 | involved_in negative regulation of DNA-templated transcription | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0046697 | involved_in decidualization | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0050790 | involved_in regulation of catalytic activity | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051287 | enables NAD binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0051901 | involved_in positive regulation of mitochondrial depolarization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060391 | involved_in positive regulation of SMAD protein signal transduction | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060545 | involved_in positive regulation of necroptotic process | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0061629 | enables RNA polymerase II-specific DNA-binding transcription factor binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0070212 | involved_in protein poly-ADP-ribosylation | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0070213 | involved_in protein auto-ADP-ribosylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0070412 | enables R-SMAD binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0071168 | involved_in protein localization to chromatin | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071294 | involved_in cellular response to zinc ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071932 | involved_in replication fork reversal | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0090734 | is_active_in site of DNA damage | 2 | IDA,IMP | Homo_sapiens(9606) | Component |
| GO:0140294 | enables NAD DNA ADP-ribosyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0140805 | enables NAD+-protein-serine ADP-ribosyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0140806 | enables NAD+- protein-aspartate ADP-ribosyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0140807 | enables NAD+-protein-glutamate ADP-ribosyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0140808 | enables NAD+-protein-tyrosine ADP-ribosyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0140815 | enables NAD+-protein-histidine ADP-ribosyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0140816 | enables NAD+-histone H2BS6 serine ADP-ribosyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0140817 | enables NAD+-histone H3S10 serine ADP-ribosyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0140822 | enables NAD+-histone H2BE35 glutamate ADP-ribosyltransferase activity | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:1900182 | involved_in positive regulation of protein localization to nucleus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1901216 | involved_in positive regulation of neuron death | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1903376 | involved_in regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1903518 | involved_in positive regulation of single strand break repair | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:1904044 | involved_in response to aldosterone | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1904178 | involved_in negative regulation of adipose tissue development | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:1904357 | involved_in negative regulation of telomere maintenance via telomere lengthening | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:1904646 | involved_in cellular response to amyloid-beta | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1904762 | involved_in positive regulation of myofibroblast differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1905051 | involved_in regulation of base-excision repair | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1905168 | involved_in positive regulation of double-strand break repair via homologous recombination | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1990090 | involved_in cellular response to nerve growth factor stimulus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1990404 | enables NAD+-protein ADP-ribosyltransferase activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:1990966 | involved_in ATP generation from poly-ADP-D-ribose | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:2000679 | involved_in positive regulation of transcription regulatory region DNA binding | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:2001170 | involved_in negative regulation of ATP biosynthetic process | 1 | IMP | Homo_sapiens(9606) | Process |