Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000141646 | Gene ID | 4089 | Accession | 6770 |
| Symbol | SMAD4 | Alias | JIP;DPC4;MADH4;MYHRS | Full Name | SMAD family member 4 |
| Status | Confidence | Length | 56652 bases | Strand | Plus strand |
| Position | 18 : 51028394 - 51085045 | RNA binding domain | N.A. | ||
| Summary | This gene encodes a member of the Smad family of signal transduction proteins. Smad proteins are phosphorylated and activated by transmembrane serine-threonine receptor kinases in response to transforming growth factor (TGF)-beta signaling. The product of this gene forms homomeric complexes and heteromeric complexes with other activated Smad proteins, which then accumulate in the nucleus and regulate the transcription of target genes. This protein binds to DNA and recognizes an 8-bp palindromic sequence (GTCTAGAC) called the Smad-binding element (SBE). The protein acts as a tumor suppressor and inhibits epithelial cell proliferation. It may also have an inhibitory effect on tumors by reducing angiogenesis and increasing blood vessel hyperpermeability. The encoded protein is a crucial component of the bone morphogenetic protein signaling pathway. The Smad proteins are subject to complex regulation by post-translational modifications. Mutations or deletions in this gene have been shown to result in pancreatic cancer, juvenile polyposis syndrome, and hereditary hemorrhagic telangiectasia syndrome. [provided by RefSeq, May 2022] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 27746021 | Direct Regulation of Alternative Splicing by SMAD3 through PCBP1 Is Essential to the Tumor-Promoting Role of TGF-β. | Veenu Tripathi | 2016-11-03 | Molecular cell |
| 20705240 | Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. | N Brandi Davis | 2010-08-13 | Molecular cell |
| 32899767 | miR-361-5p Mediates SMAD4 to Promote Porcine Granulosa Cell Apoptosis through VEGFA. | Mengnan Ma | 2020-09-04 | Biomolecules |
| 35794621 | Long noncoding RNA Smyca coactivates TGF-β/Smad and Myc pathways to drive tumor progression. | Hsin-Yi Chen | 2022-07-06 | Journal of hematology & oncology |
| 29149681 | Mechanistic insight into contextual TGF-β signaling. | E Ying Zhang | 2018-04-01 | Current opinion in cell biology |
| 30185897 | Malignant Transformation of Human Bronchial Epithelial Cells Induced by Arsenic through STAT3/miR-301a/SMAD4 Loop. | Mingtian Zhong | 2018-09-05 | Scientific reports |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| SMAD4-206 | ENST00000588860 | 8474 | 552aa | ENSP00000465878 |
| SMAD4-202 | ENST00000398417 | 8495 | 552aa | ENSP00000381452 |
| SMAD4-207 | ENST00000589076 | 8538 | 552aa | ENSP00000466934 |
| SMAD4-209 | ENST00000589941 | 8941 | 552aa | ENSP00000465874 |
| SMAD4-201 | ENST00000342988 | 8772 | 552aa | ENSP00000341551 |
| SMAD4-210 | ENST00000590061 | 8357 | 552aa | ENSP00000464772 |
| SMAD4-215 | ENST00000593223 | 4980 | 493aa | ENSP00000466118 |
| SMAD4-214 | ENST00000592911 | 475 | No protein | - |
| SMAD4-216 | ENST00000611848 | 2906 | 486aa | ENSP00000478613 |
| SMAD4-205 | ENST00000588745 | 1371 | 456aa | ENSP00000464901 |
| SMAD4-213 | ENST00000592186 | 1306 | 320aa | ENSP00000468611 |
| SMAD4-208 | ENST00000589706 | 563 | No protein | - |
| SMAD4-203 | ENST00000585448 | 459 | No protein | - |
| SMAD4-212 | ENST00000591126 | 4970 | No protein | - |
| SMAD4-217 | ENST00000684953 | 5164 | No protein | - |
| SMAD4-220 | ENST00000688307 | 1146 | No protein | - |
| SMAD4-222 | ENST00000688903 | 1963 | No protein | - |
| SMAD4-223 | ENST00000690892 | 1223 | No protein | - |
| SMAD4-219 | ENST00000685232 | 2883 | No protein | - |
| SMAD4-221 | ENST00000688574 | 8307 | No protein | - |
| SMAD4-218 | ENST00000685090 | 4891 | No protein | - |
| SMAD4-224 | ENST00000691124 | 5780 | No protein | - |
| SMAD4-211 | ENST00000590499 | 545 | No protein | - |
| SMAD4-204 | ENST00000586253 | 784 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 54565 | Beta-Globulins | 5.9390000E-005 | - |
| 54734 | Ferritins | 1.0860000E-005 | - |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
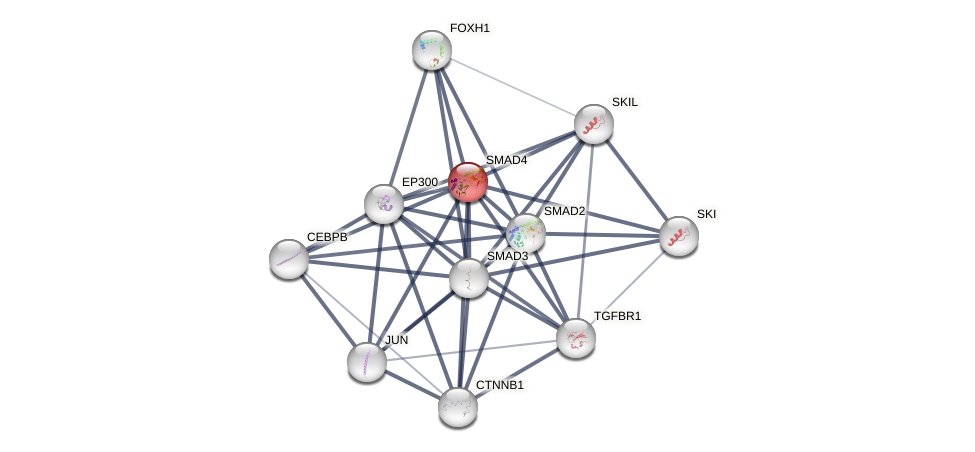
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000141646 | homo_sapiens | ENSP00000369154 | 54.6039 | 39.6146 | homo_sapiens | ENSP00000341551 | 46.1957 | 33.5145 |
| ENSG00000141646 | homo_sapiens | ENSP00000262160 | 53.9615 | 40.6852 | homo_sapiens | ENSP00000341551 | 45.6522 | 34.4203 |
| ENSG00000141646 | homo_sapiens | ENSP00000332973 | 60.2353 | 44.4706 | homo_sapiens | ENSP00000341551 | 46.3768 | 34.2391 |
| ENSG00000141646 | homo_sapiens | ENSP00000305769 | 55.0538 | 39.3548 | homo_sapiens | ENSP00000341551 | 46.3768 | 33.1522 |
| ENSG00000141646 | homo_sapiens | ENSP00000441954 | 56.129 | 40.6452 | homo_sapiens | ENSP00000341551 | 47.2826 | 34.2391 |
| ENSG00000141646 | homo_sapiens | ENSP00000288840 | 36.8952 | 25.6048 | homo_sapiens | ENSP00000341551 | 33.1522 | 23.0072 |
| ENSG00000141646 | homo_sapiens | ENSP00000262158 | 41.3146 | 24.1784 | homo_sapiens | ENSP00000341551 | 31.8841 | 18.6594 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 100 | nomascus_leucogenys | ENSNLEP00000014415 | 100 | 100 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 100 | pan_paniscus | ENSPPAP00000009478 | 100 | 100 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 100 | pongo_abelii | ENSPPYP00000010297 | 100 | 100 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 100 | pan_troglodytes | ENSPTRP00000017047 | 100 | 100 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 100 | gorilla_gorilla | ENSGGOP00000013332 | 100 | 100 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.5507 | 97.4638 | ornithorhynchus_anatinus | ENSOANP00000012901 | 98.5507 | 97.4638 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.5942 | 85.1449 | sarcophilus_harrisii | ENSSHAP00000028104 | 98.3539 | 96.7078 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 78.9855 | 78.2609 | notamacropus_eugenii | ENSMEUP00000001667 | 79.1289 | 78.4029 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 82.971 | 82.6087 | choloepus_hoffmanni | ENSCHOP00000000766 | 98.4946 | 98.0645 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.0942 | 98.7319 | dasypus_novemcinctus | ENSDNOP00000003926 | 99.0942 | 98.7319 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 90.0362 | 89.1304 | erinaceus_europaeus | ENSEEUP00000001427 | 90.1996 | 89.2922 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.913 | 97.8261 | echinops_telfairi | ENSETEP00000006297 | 98.913 | 97.8261 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.1884 | 98.0072 | callithrix_jacchus | ENSCJAP00000056533 | 89.5868 | 89.4215 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 100 | cercocebus_atys | ENSCATP00000036741 | 100 | 100 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 100 | macaca_fascicularis | ENSMFAP00000032134 | 100 | 100 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 100 | macaca_mulatta | ENSMMUP00000024106 | 100 | 100 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 100 | macaca_nemestrina | ENSMNEP00000013301 | 100 | 100 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 100 | papio_anubis | ENSPANP00000006643 | 100 | 100 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 100 | mandrillus_leucophaeus | ENSMLEP00000007644 | 100 | 100 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 72.6449 | 72.2826 | tursiops_truncatus | ENSTTRP00000011160 | 98.5258 | 98.0344 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 98.913 | vulpes_vulpes | ENSVVUP00000019738 | 99.2754 | 98.913 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.913 | 98.1884 | mus_spretus | MGP_SPRETEiJ_P0049055 | 99.0926 | 98.3666 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 99.0942 | equus_asinus | ENSEASP00005042800 | 99.2754 | 99.0942 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 92.2101 | 92.029 | capra_hircus | ENSCHIP00000020573 | 99.2203 | 99.0253 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 99.0942 | loxodonta_africana | ENSLAFP00000010529 | 99.2754 | 99.0942 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | panthera_leo | ENSPLOP00000016998 | 99.4565 | 99.2754 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.0942 | ailuropoda_melanoleuca | ENSAMEP00000003072 | 99.4565 | 99.0942 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | bos_indicus_hybrid | ENSBIXP00005022794 | 99.2767 | 99.0958 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.913 | 98.7319 | oryctolagus_cuniculus | ENSOCUP00000003411 | 98.913 | 98.7319 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 99.0942 | equus_caballus | ENSECAP00000042762 | 99.2754 | 99.0942 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.913 | 98.1884 | canis_lupus_familiaris | ENSCAFP00845004513 | 99.2727 | 98.5455 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | panthera_pardus | ENSPPRP00000018849 | 99.4565 | 99.2754 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | marmota_marmota_marmota | ENSMMMP00000006303 | 99.4565 | 99.2754 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 98.7319 | balaenoptera_musculus | ENSBMSP00010013498 | 99.0958 | 98.5533 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | cavia_porcellus | ENSCPOP00000005827 | 99.4565 | 99.2754 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | felis_catus | ENSFCAP00000031311 | 99.4565 | 99.2754 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.0942 | ursus_americanus | ENSUAMP00000018392 | 99.4565 | 99.0942 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 95.471 | 94.3841 | monodelphis_domestica | ENSMODP00000037121 | 98.6891 | 97.5655 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | bison_bison_bison | ENSBBBP00000016297 | 99.2767 | 99.0958 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.0942 | monodon_monoceros | ENSMMNP00015000620 | 99.2767 | 98.915 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 98.913 | sus_scrofa | ENSSSCP00000004879 | 99.2754 | 98.913 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.0942 | microcebus_murinus | ENSMICP00000038516 | 99.4565 | 99.0942 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.0942 | octodon_degus | ENSODEP00000025235 | 99.4565 | 99.0942 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 97.1014 | 96.7391 | jaculus_jaculus | ENSJJAP00000004333 | 99.0758 | 98.7061 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | ovis_aries_rambouillet | ENSOARP00020020393 | 99.2767 | 99.0958 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.1884 | 97.6449 | ursus_maritimus | ENSUMAP00000003792 | 99.2674 | 98.7179 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.7319 | 98.1884 | ochotona_princeps | ENSOPRP00000012406 | 98.7319 | 98.1884 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.0942 | delphinapterus_leucas | ENSDLEP00000005877 | 99.2767 | 98.915 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.0942 | physeter_catodon | ENSPCTP00005019284 | 99.2767 | 98.915 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.913 | 98.3696 | mus_caroli | MGP_CAROLIEiJ_P0047101 | 99.0926 | 98.5481 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.913 | 98.3696 | mus_musculus | ENSMUSP00000025393 | 99.0926 | 98.5481 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 89.4928 | 86.9565 | dipodomys_ordii | ENSDORP00000008535 | 93.5606 | 90.9091 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.0942 | mustela_putorius_furo | ENSMPUP00000008705 | 99.4565 | 99.0942 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.8188 | 99.8188 | aotus_nancymaae | ENSANAP00000014952 | 99.8188 | 99.8188 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 100 | saimiri_boliviensis_boliviensis | ENSSBOP00000026561 | 100 | 100 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.1884 | 98.0072 | vicugna_pacos | ENSVPAP00000006278 | 98.9051 | 98.7226 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.8188 | 99.8188 | chlorocebus_sabaeus | ENSCSAP00000014115 | 99.8188 | 99.8188 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 77.1739 | 76.087 | tupaia_belangeri | ENSTBEP00000012234 | 77.5956 | 76.5027 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.0942 | 98.913 | mesocricetus_auratus | ENSMAUP00000008260 | 99.0942 | 98.913 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.0942 | phocoena_sinus | ENSPSNP00000010840 | 99.2767 | 98.915 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | camelus_dromedarius | ENSCDRP00005012694 | 99.4565 | 99.2754 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.0942 | 98.7319 | carlito_syrichta | ENSTSYP00000009006 | 99.274 | 98.9111 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | panthera_tigris_altaica | ENSPTIP00000002665 | 99.4565 | 99.2754 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.913 | 98.7319 | rattus_norvegicus | ENSRNOP00000074882 | 98.913 | 98.7319 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 98.5507 | prolemur_simus | ENSPSMP00000029655 | 99.2754 | 98.5507 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.0942 | 98.913 | microtus_ochrogaster | ENSMOCP00000015787 | 99.0942 | 98.913 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.058 | 83.5145 | sorex_araneus | ENSSARP00000012542 | 99.1453 | 98.5043 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.0942 | 98.913 | peromyscus_maniculatus_bairdii | ENSPEMP00000013693 | 99.0942 | 98.913 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | bos_mutus | ENSBMUP00000032268 | 99.2767 | 99.0958 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.913 | 98.3696 | mus_spicilegus | ENSMSIP00000012826 | 99.0926 | 98.5481 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.0942 | 98.913 | cricetulus_griseus_chok1gshd | ENSCGRP00001024277 | 99.0942 | 98.913 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.3261 | 85.1449 | pteropus_vampyrus | ENSPVAP00000004034 | 85.3261 | 85.1449 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 99.0942 | myotis_lucifugus | ENSMLUP00000012691 | 98.917 | 98.7365 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.5507 | 97.2826 | vombatus_ursinus | ENSVURP00010001387 | 98.5507 | 97.2826 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.0942 | rhinolophus_ferrumequinum | ENSRFEP00010025005 | 99.4565 | 99.0942 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.0942 | neovison_vison | ENSNVIP00000003495 | 99.4565 | 99.0942 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | bos_taurus | ENSBTAP00000009081 | 99.2767 | 99.0958 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 99.0942 | heterocephalus_glaber_female | ENSHGLP00000007949 | 99.2754 | 99.0942 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 99.6377 | rhinopithecus_bieti | ENSRBIP00000018874 | 100 | 99.6377 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 98.913 | canis_lupus_dingo | ENSCAFP00020027598 | 99.2754 | 98.913 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 99.0942 | sciurus_vulgaris | ENSSVLP00005029879 | 99.2754 | 99.0942 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.5507 | 97.2826 | phascolarctos_cinereus | ENSPCIP00000019556 | 98.5507 | 97.2826 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 75.3623 | 74.8188 | procavia_capensis | ENSPCAP00000008572 | 93.4831 | 92.809 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 98.913 | nannospalax_galili | ENSNGAP00000002152 | 99.2754 | 98.913 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 95.8333 | 95.6522 | bos_grunniens | ENSBGRP00000039068 | 97.4217 | 97.2376 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | chinchilla_lanigera | ENSCLAP00000013454 | 99.4565 | 99.2754 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | cervus_hanglu_yarkandensis | ENSCHYP00000039168 | 99.2767 | 99.0958 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 98.913 | catagonus_wagneri | ENSCWAP00000023146 | 99.2754 | 98.913 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | ictidomys_tridecemlineatus | ENSSTOP00000016654 | 99.4565 | 99.2754 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.2754 | 98.7319 | otolemur_garnettii | ENSOGAP00000010810 | 99.2754 | 98.7319 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.9638 | 84.9638 | cebus_imitator | ENSCCAP00000030667 | 100 | 100 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 94.7464 | 94.5652 | urocitellus_parryii | ENSUPAP00010019327 | 94.7464 | 94.5652 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.2754 | moschus_moschiferus | ENSMMSP00000007203 | 99.2767 | 99.0958 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 100 | 99.6377 | rhinopithecus_roxellana | ENSRROP00000043301 | 100 | 99.6377 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.3696 | 98.1884 | mus_pahari | MGP_PahariEiJ_P0039673 | 98.5481 | 98.3666 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 99.4565 | 99.0942 | propithecus_coquereli | ENSPCOP00000024999 | 99.4565 | 99.0942 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 26.8116 | 15.7609 | caenorhabditis_elegans | F37D6.6.1 | 35.6627 | 20.9639 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 26.4493 | 15.3986 | caenorhabditis_elegans | F01G10.8b.1 | 44.1088 | 25.6798 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 25.7246 | 15.7609 | drosophila_melanogaster | FBpp0428153 | 21.3534 | 13.0827 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 25.1812 | 14.6739 | ciona_intestinalis | ENSCINP00000018319 | 38.5042 | 22.4377 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 28.0797 | 19.9275 | ciona_intestinalis | ENSCINP00000012287 | 45.0581 | 31.9767 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 41.4855 | 35.6884 | petromyzon_marinus | ENSPMAP00000009249 | 75.082 | 64.5902 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 26.4493 | 15.942 | eptatretus_burgeri | ENSEBUP00000021174 | 30.9322 | 18.6441 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 92.029 | 86.9565 | callorhinchus_milii | ENSCMIP00000031842 | 91.8626 | 86.7993 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 69.9275 | 65.7609 | latimeria_chalumnae | ENSLACP00000009312 | 93.6893 | 88.1068 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.7754 | 80.9783 | lepisosteus_oculatus | ENSLOCP00000015848 | 87.5686 | 81.7185 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.8696 | 79.529 | clupea_harengus | ENSCHAP00000061349 | 88.2682 | 81.7505 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 92.5725 | 86.7754 | clupea_harengus | ENSCHAP00000016001 | 92.5725 | 86.7754 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 52.3551 | 46.0145 | danio_rerio | ENSDARP00000099850 | 87.0482 | 76.506 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 92.9348 | 87.6812 | danio_rerio | ENSDARP00000097763 | 93.7843 | 88.4826 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 92.2101 | 86.5942 | carassius_auratus | ENSCARP00000095408 | 93.053 | 87.3857 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 92.5725 | 86.7754 | carassius_auratus | ENSCARP00000012161 | 93.4186 | 87.5686 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 75.7246 | 69.3841 | carassius_auratus | ENSCARP00000001331 | 87.2651 | 79.9582 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 83.6957 | 76.9928 | carassius_auratus | ENSCARP00000010492 | 87.1698 | 80.1887 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 92.2101 | 86.5942 | carassius_auratus | ENSCARP00000084540 | 93.053 | 87.3857 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 92.7536 | 86.7754 | carassius_auratus | ENSCARP00000005317 | 93.6015 | 87.5686 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 79.3478 | 73.7319 | astyanax_mexicanus | ENSAMXP00000028486 | 88.8438 | 82.5558 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 91.1232 | 85.8696 | astyanax_mexicanus | ENSAMXP00000019787 | 92.4632 | 87.1324 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 79.529 | 72.8261 | ictalurus_punctatus | ENSIPUP00000035179 | 83.9388 | 76.8642 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 91.6667 | 86.5942 | ictalurus_punctatus | ENSIPUP00000007352 | 92.674 | 87.5458 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 88.4058 | 82.4275 | esox_lucius | ENSELUP00000020999 | 90.7063 | 84.5725 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 89.6739 | 83.5145 | esox_lucius | ENSELUP00000024614 | 82.7759 | 77.0903 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 92.029 | 86.2319 | electrophorus_electricus | ENSEEEP00000020048 | 93.211 | 87.3394 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 83.8768 | 76.2681 | electrophorus_electricus | ENSEEEP00000016285 | 88.3588 | 80.3435 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 82.971 | 77.8986 | oncorhynchus_kisutch | ENSOKIP00005089085 | 79.5139 | 74.6528 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 88.587 | 82.6087 | oncorhynchus_kisutch | ENSOKIP00005054839 | 88.9091 | 82.9091 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 89.6739 | 83.3333 | oncorhynchus_kisutch | ENSOKIP00005050460 | 90.1639 | 83.7887 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.5 | 82.4275 | oncorhynchus_kisutch | ENSOKIP00005005862 | 88.1387 | 83.0292 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.413 | 80.9783 | oncorhynchus_mykiss | ENSOMYP00000142378 | 86.2568 | 80.8318 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.6014 | 79.3478 | oncorhynchus_mykiss | ENSOMYP00000099836 | 85.3748 | 80.0731 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 88.7681 | 82.4275 | oncorhynchus_mykiss | ENSOMYP00000061272 | 89.4161 | 83.0292 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 89.4928 | 82.6087 | oncorhynchus_mykiss | ENSOMYP00000058974 | 89.3309 | 82.4593 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.6014 | 79.7101 | salmo_salar | ENSSSAP00000101532 | 91.5686 | 86.2745 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 89.6739 | 82.7899 | salmo_salar | ENSSSAP00000001454 | 89.3502 | 82.491 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 89.1304 | 82.971 | salmo_salar | ENSSSAP00000070778 | 89.781 | 83.5766 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.1377 | 82.0652 | salmo_salar | ENSSSAP00000104615 | 87.1377 | 82.0652 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.1377 | 82.0652 | salmo_trutta | ENSSTUP00000077093 | 89.2393 | 84.0445 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.6884 | 80.4348 | salmo_trutta | ENSSTUP00000031260 | 86.4717 | 81.17 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 89.1304 | 82.971 | salmo_trutta | ENSSTUP00000035981 | 89.781 | 83.5766 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 89.8551 | 82.7899 | salmo_trutta | ENSSTUP00000094230 | 89.5307 | 82.491 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.8623 | 82.2464 | gadus_morhua | ENSGMOP00000048012 | 89.6488 | 83.9187 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 27.5362 | 25.5435 | fundulus_heteroclitus | ENSFHEP00000029923 | 77.551 | 71.9388 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 61.0507 | 55.2536 | fundulus_heteroclitus | ENSFHEP00000032911 | 85.7506 | 77.6081 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 74.2754 | 70.8333 | poecilia_reticulata | ENSPREP00000027467 | 93.3941 | 89.0661 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.2391 | 80.0725 | poecilia_reticulata | ENSPREP00000020499 | 92.4453 | 87.8728 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.413 | 81.8841 | xiphophorus_maculatus | ENSXMAP00000031130 | 93.1641 | 88.2812 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.4203 | 80.0725 | xiphophorus_maculatus | ENSXMAP00000005074 | 92.6441 | 87.8728 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 82.971 | 77.1739 | oryzias_latipes | ENSORLP00000045202 | 86.7424 | 80.6818 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.3188 | 81.5217 | oryzias_latipes | ENSORLP00000006328 | 94.5098 | 88.2353 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.6014 | 77.3551 | cyclopterus_lumpus | ENSCLMP00005028472 | 88.1132 | 80.566 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.7754 | 82.4275 | cyclopterus_lumpus | ENSCLMP00005004514 | 92.1154 | 87.5 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.5942 | 80.9783 | oreochromis_niloticus | ENSONIP00000050523 | 90.8745 | 84.981 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 88.0435 | 82.4275 | oreochromis_niloticus | ENSONIP00000072817 | 88.3636 | 82.7273 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.9638 | 80.4348 | haplochromis_burtoni | ENSHBUP00000019491 | 93.2406 | 88.2704 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.0507 | 81.5217 | haplochromis_burtoni | ENSHBUP00000008979 | 93.1373 | 88.2353 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.9638 | 80.4348 | astatotilapia_calliptera | ENSACLP00000021363 | 93.2406 | 88.2704 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.0507 | 81.5217 | astatotilapia_calliptera | ENSACLP00000025950 | 93.1373 | 88.2353 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.8623 | 83.3333 | sparus_aurata | ENSSAUP00010014730 | 93.4489 | 88.632 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.4203 | 79.529 | sparus_aurata | ENSSAUP00010059920 | 91.0156 | 85.7422 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.7754 | 81.5217 | lates_calcarifer | ENSLCAP00010013760 | 91.4122 | 85.8779 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 88.9493 | 83.8768 | lates_calcarifer | ENSLCAP00010001596 | 92.467 | 87.194 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 95.2899 | 91.8478 | xenopus_tropicalis | ENSXETP00000048372 | 95.2899 | 91.8478 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.1884 | 96.558 | chrysemys_picta_bellii | ENSCPBP00000036916 | 98.3666 | 96.7332 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 97.4638 | 95.1087 | sphenodon_punctatus | ENSSPUP00000022823 | 97.4638 | 95.1087 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 97.6449 | 94.9275 | laticauda_laticaudata | ENSLLTP00000001203 | 97.6449 | 94.9275 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 97.6449 | 94.9275 | notechis_scutatus | ENSNSUP00000017870 | 97.6449 | 94.9275 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 97.6449 | 94.9275 | pseudonaja_textilis | ENSPTXP00000025616 | 97.6449 | 94.9275 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 97.8261 | 95.8333 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000003446 | 98.0036 | 96.0073 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 78.8043 | 75 | gallus_gallus | ENSGALP00010012877 | 90.4366 | 86.0707 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 91.4855 | 87.6812 | gallus_gallus | ENSGALP00010016770 | 77.3354 | 74.1194 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 26.9928 | 26.2681 | meleagris_gallopavo | ENSMGAP00000021285 | 99.3333 | 96.6667 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 91.4855 | 89.6739 | serinus_canaria | ENSSCAP00000002622 | 98.8258 | 96.8689 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 38.4058 | 35.6884 | parus_major | ENSPMJP00000002454 | 90.2128 | 83.8298 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 90.2174 | 86.0507 | anolis_carolinensis | ENSACAP00000026900 | 83.9798 | 80.1012 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.2319 | 81.7029 | neolamprologus_brichardi | ENSNBRP00000016107 | 93.3333 | 88.4314 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.5072 | 80.6159 | neolamprologus_brichardi | ENSNBRP00000005979 | 92.0078 | 86.7446 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 83.5145 | 79.7101 | naja_naja | ENSNNAP00000019986 | 94.4672 | 90.1639 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.1884 | 96.3768 | podarcis_muralis | ENSPMRP00000015240 | 98.1884 | 96.3768 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 91.3043 | 89.3116 | geospiza_fortis | ENSGFOP00000006917 | 98.6301 | 96.4775 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 88.7681 | 84.9638 | taeniopygia_guttata | ENSTGUP00000032640 | 95.3307 | 91.2451 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 93.1159 | 87.5 | erpetoichthys_calabaricus | ENSECRP00000006242 | 93.6248 | 87.9781 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 78.442 | 74.8188 | kryptolebias_marmoratus | ENSKMAP00000028560 | 94.7484 | 90.372 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.7826 | 79.8913 | kryptolebias_marmoratus | ENSKMAP00000013171 | 93.0417 | 87.674 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 97.6449 | 95.6522 | salvator_merianae | ENSSMRP00000021246 | 97.8221 | 95.8258 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.6812 | 82.0652 | oryzias_javanicus | ENSOJAP00000046672 | 85.968 | 80.4618 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.2391 | 78.9855 | oryzias_javanicus | ENSOJAP00000008797 | 91.1765 | 85.4902 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.8696 | 80.9783 | amphilophus_citrinellus | ENSACIP00000023940 | 91.1538 | 85.9615 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 81.3406 | 77.3551 | amphilophus_citrinellus | ENSACIP00000007188 | 93.737 | 89.1441 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 88.2246 | 82.6087 | dicentrarchus_labrax | ENSDLAP00005046845 | 90.689 | 84.9162 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.9638 | 80.0725 | dicentrarchus_labrax | ENSDLAP00005082836 | 89.1635 | 84.0304 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.1377 | 82.6087 | amphiprion_percula | ENSAPEP00000004950 | 94.685 | 89.7638 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.9638 | 80.2536 | amphiprion_percula | ENSAPEP00000017835 | 93.2406 | 88.0716 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 69.7464 | 64.1304 | oryzias_sinensis | ENSOSIP00000006389 | 92.7711 | 85.3012 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.3261 | 78.9855 | oryzias_sinensis | ENSOSIP00000008590 | 86.9004 | 80.4428 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.1884 | 96.0145 | pelodiscus_sinensis | ENSPSIP00000016512 | 98.1884 | 96.0145 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.1884 | 96.558 | gopherus_evgoodei | ENSGEVP00005027739 | 98.3666 | 96.7332 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.1884 | 96.558 | chelonoidis_abingdonii | ENSCABP00000017324 | 98.3666 | 96.7332 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.3261 | 80.6159 | seriola_dumerili | ENSSDUP00000007227 | 93.6382 | 88.4692 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 88.0435 | 83.5145 | seriola_dumerili | ENSSDUP00000030892 | 95.2941 | 90.3922 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.2319 | 79.529 | cottoperca_gobio | ENSCGOP00000029950 | 89.8113 | 82.8302 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.5072 | 80.6159 | cottoperca_gobio | ENSCGOP00000028769 | 92.0078 | 86.7446 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.0072 | 96.0145 | struthio_camelus_australis | ENSSCUP00000002001 | 94.9123 | 92.9825 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 97.8261 | 95.8333 | anser_brachyrhynchus | ENSABRP00000024877 | 98.0036 | 96.0073 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 83.3333 | 75.9058 | gasterosteus_aculeatus | ENSGACP00000022267 | 87.4525 | 79.6578 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 88.0435 | 83.1522 | gasterosteus_aculeatus | ENSGACP00000009949 | 88.6861 | 83.7591 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.3261 | 80.9783 | cynoglossus_semilaevis | ENSCSEP00000026341 | 93.6382 | 88.8668 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.2319 | 80.4348 | cynoglossus_semilaevis | ENSCSEP00000010634 | 93.3333 | 87.0588 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.5942 | 81.8841 | poecilia_formosa | ENSPFOP00000024750 | 93.7255 | 88.6274 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.4203 | 80.0725 | poecilia_formosa | ENSPFOP00000019933 | 92.6441 | 87.8728 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.5942 | 81.1594 | myripristis_murdjan | ENSMMDP00005033818 | 90.1887 | 84.5283 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 88.2246 | 82.4275 | myripristis_murdjan | ENSMMDP00005055330 | 90.1852 | 84.2593 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 80.4348 | 75.5435 | sander_lucioperca | ENSSLUP00000038549 | 91.9255 | 86.3354 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.1377 | 82.6087 | sander_lucioperca | ENSSLUP00000016424 | 90.7547 | 86.0377 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 97.4638 | 95.471 | strigops_habroptila | ENSSHBP00005000015 | 97.6407 | 95.6443 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 91.4855 | 89.6739 | strigops_habroptila | ENSSHBP00005013289 | 98.8258 | 96.8689 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.9638 | 80.2536 | amphiprion_ocellaris | ENSAOCP00000016055 | 93.2406 | 88.0716 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.5 | 82.6087 | amphiprion_ocellaris | ENSAOCP00000016426 | 94.7059 | 89.4118 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.1884 | 96.558 | terrapene_carolina_triunguis | ENSTMTP00000000092 | 98.3666 | 96.7332 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 89.8551 | 84.058 | hucho_hucho | ENSHHUP00000033303 | 89.8551 | 84.058 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.413 | 81.3406 | hucho_hucho | ENSHHUP00000026360 | 89.4934 | 84.2402 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 90.2174 | 83.8768 | hucho_hucho | ENSHHUP00000068309 | 90.0543 | 83.7251 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 83.5145 | 79.529 | hucho_hucho | ENSHHUP00000082227 | 89.6887 | 85.4086 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.2319 | 80.6159 | betta_splendens | ENSBSLP00000011769 | 92.607 | 86.5759 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.7754 | 80.9783 | betta_splendens | ENSBSLP00000018322 | 88.7037 | 82.7778 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 91.8478 | 86.9565 | pygocentrus_nattereri | ENSPNAP00000013918 | 92.6874 | 87.7514 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 88.2246 | 83.5145 | anabas_testudineus | ENSATEP00000021861 | 93.8343 | 88.8247 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.6884 | 80.7971 | anabas_testudineus | ENSATEP00000067701 | 90.2672 | 85.1145 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 24.6377 | 13.9493 | ciona_savignyi | ENSCSAVP00000018932 | 37.7778 | 21.3889 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.3261 | 80.6159 | seriola_lalandi_dorsalis | ENSSLDP00000014268 | 93.6382 | 88.4692 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 88.0435 | 83.5145 | seriola_lalandi_dorsalis | ENSSLDP00000032438 | 95.2941 | 90.3922 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.6884 | 80.2536 | labrus_bergylta | ENSLBEP00000009797 | 90.7869 | 85.0288 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.6014 | 80.0725 | labrus_bergylta | ENSLBEP00000023864 | 92.8429 | 87.8728 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.9638 | 80.4348 | pundamilia_nyererei | ENSPNYP00000010707 | 93.2406 | 88.2704 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.0507 | 81.5217 | pundamilia_nyererei | ENSPNYP00000013790 | 93.1373 | 88.2353 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.5 | 82.4275 | mastacembelus_armatus | ENSMAMP00000001989 | 90.9604 | 85.6874 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.8696 | 80.7971 | mastacembelus_armatus | ENSMAMP00000048564 | 89.2655 | 83.9925 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.9638 | 80.0725 | stegastes_partitus | ENSSPAP00000005949 | 93.2406 | 87.8728 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.3188 | 82.2464 | stegastes_partitus | ENSSPAP00000003348 | 91.2879 | 85.9848 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.4203 | 79.1667 | oncorhynchus_tshawytscha | ENSOTSP00005009763 | 87.4296 | 81.9887 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 89.1304 | 82.6087 | oncorhynchus_tshawytscha | ENSOTSP00005052633 | 87.5445 | 81.1388 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 82.971 | 78.0797 | oncorhynchus_tshawytscha | ENSOTSP00005089484 | 75.8278 | 71.3576 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 89.3116 | 82.4275 | oncorhynchus_tshawytscha | ENSOTSP00005091052 | 89.1501 | 82.2785 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.9638 | 80.2536 | acanthochromis_polyacanthus | ENSAPOP00000008331 | 93.2406 | 88.0716 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.6812 | 82.7899 | acanthochromis_polyacanthus | ENSAPOP00000020671 | 94.902 | 89.6078 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 98.0072 | 96.0145 | aquila_chrysaetos_chrysaetos | ENSACCP00020013966 | 98.1851 | 96.1888 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.6014 | 79.529 | nothobranchius_furzeri | ENSNFUP00015031932 | 88.1132 | 82.8302 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.6812 | 81.8841 | nothobranchius_furzeri | ENSNFUP00015036278 | 90.8068 | 84.803 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.3188 | 82.6087 | cyprinodon_variegatus | ENSCVAP00000004754 | 93.4109 | 88.3721 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.7826 | 79.8913 | cyprinodon_variegatus | ENSCVAP00000018520 | 93.0417 | 87.674 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.7826 | 78.6232 | denticeps_clupeoides | ENSDCDP00000008359 | 89.313 | 82.8244 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 81.1594 | 73.1884 | denticeps_clupeoides | ENSDCDP00000016650 | 74.6667 | 67.3333 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.6884 | 80.4348 | larimichthys_crocea | ENSLCRP00005027948 | 90.0952 | 84.5714 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 88.0435 | 83.3333 | larimichthys_crocea | ENSLCRP00005034801 | 89.6679 | 84.8708 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.6884 | 80.4348 | takifugu_rubripes | ENSTRUP00000048686 | 90.7869 | 85.2207 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 32.0652 | 28.442 | takifugu_rubripes | ENSTRUP00000076792 | 75 | 66.5254 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 73.3696 | 67.5725 | takifugu_rubripes | ENSTRUP00000053126 | 72.711 | 66.9659 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 83.6957 | 79.529 | hippocampus_comes | ENSHCOP00000015228 | 91.8489 | 87.2763 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.2391 | 78.9855 | hippocampus_comes | ENSHCOP00000022820 | 91.1765 | 85.4902 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 92.9348 | 86.9565 | cyprinus_carpio_carpio | ENSCCRP00000045211 | 93.7843 | 87.7514 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 70.2899 | 61.2319 | cyprinus_carpio_carpio | ENSCCRP00000037883 | 85.2747 | 74.2857 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 92.2101 | 86.0507 | cyprinus_carpio_carpio | ENSCCRP00000156841 | 92.8832 | 86.6788 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.1449 | 77.3551 | cyprinus_carpio_carpio | ENSCCRP00000106773 | 86.0806 | 78.2051 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 74.4565 | 71.3768 | coturnix_japonica | ENSCJPP00005001081 | 86.5263 | 82.9474 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.4203 | 80.0725 | poecilia_latipinna | ENSPLAP00000003577 | 92.6441 | 87.8728 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.5942 | 82.0652 | poecilia_latipinna | ENSPLAP00000019866 | 93.7255 | 88.8235 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 80.2536 | 74.4565 | sinocyclocheilus_grahami | ENSSGRP00000022575 | 88.7776 | 82.3647 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 72.4638 | 66.1232 | sinocyclocheilus_grahami | ENSSGRP00000010396 | 86.2069 | 78.6638 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 69.5652 | 61.0507 | sinocyclocheilus_grahami | ENSSGRP00000029869 | 79.3388 | 69.6281 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 92.5725 | 87.5 | sinocyclocheilus_grahami | ENSSGRP00000007947 | 92.9091 | 87.8182 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.0507 | 81.5217 | maylandia_zebra | ENSMZEP00005006313 | 93.1373 | 88.2353 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 84.9638 | 80.4348 | maylandia_zebra | ENSMZEP00005024930 | 93.2406 | 88.2704 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.3261 | 79.8913 | tetraodon_nigroviridis | ENSTNIP00000005773 | 92.3529 | 86.4706 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 85.5072 | 80.6159 | tetraodon_nigroviridis | ENSTNIP00000011908 | 88.5553 | 83.4897 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 91.8478 | 86.0507 | paramormyrops_kingsleyae | ENSPKIP00000039533 | 93.0275 | 87.156 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 89.3116 | 84.058 | scleropages_formosus | ENSSFOP00015015148 | 89.4737 | 84.2105 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 94.3841 | 90.3986 | leptobrachium_leishanense | ENSLLEP00000006190 | 89.8276 | 86.0345 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 86.9565 | 82.971 | scophthalmus_maximus | ENSSMAP00000001661 | 93.75 | 89.4531 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 76.2681 | 71.3768 | scophthalmus_maximus | ENSSMAP00000053749 | 78.9869 | 73.9212 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 83.1522 | 78.2609 | oryzias_melastigma | ENSOMEP00000023058 | 93.1034 | 87.6268 |
| ENSG00000141646 | homo_sapiens | ENSP00000341551 | 87.6812 | 81.8841 | oryzias_melastigma | ENSOMEP00000021474 | 94.902 | 88.6274 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000122 | involved_in negative regulation of transcription by RNA polymerase II | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0000785 | part_of chromatin | 2 | IDA,ISA | Homo_sapiens(9606) | Component |
| GO:0000976 | enables transcription cis-regulatory region binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0000978 | enables RNA polymerase II cis-regulatory region sequence-specific DNA binding | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0000981 | enables DNA-binding transcription factor activity, RNA polymerase II-specific | 2 | IBA,ISA | Homo_sapiens(9606) | Function |
| GO:0001223 | enables transcription coactivator binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0001228 | enables DNA-binding transcription activator activity, RNA polymerase II-specific | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0001541 | involved_in ovarian follicle development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001649 | involved_in osteoblast differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001658 | involved_in branching involved in ureteric bud morphogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001666 | involved_in response to hypoxia | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0001701 | involved_in in utero embryonic development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001702 | involved_in gastrulation with mouth forming second | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001837 | involved_in epithelial to mesenchymal transition | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0003148 | involved_in outflow tract septum morphogenesis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0003161 | involved_in cardiac conduction system development | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0003190 | involved_in atrioventricular valve formation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0003198 | involved_in epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0003220 | involved_in left ventricular cardiac muscle tissue morphogenesis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0003251 | involved_in positive regulation of cell proliferation involved in heart valve morphogenesis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0003360 | involved_in brainstem development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0003677 | contributes_to DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003682 | enables chromatin binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0003700 | contributes_to DNA-binding transcription factor activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005518 | enables collagen binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005667 | part_of transcription regulator complex | 2 | IDA,IPI | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005813 | located_in centrosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0006351 | involved_in DNA-templated transcription | 2 | IDA,NAS | Homo_sapiens(9606) | Process |
| GO:0006357 | involved_in regulation of transcription by RNA polymerase II | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0006366 | involved_in transcription by RNA polymerase II | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006879 | involved_in intracellular iron ion homeostasis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0007179 | involved_in transforming growth factor beta receptor signaling pathway | 4 | IBA,IDA,IMP,ISS | Homo_sapiens(9606) | Process |
| GO:0007283 | involved_in spermatogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007338 | involved_in single fertilization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007411 | involved_in axon guidance | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008283 | involved_in cell population proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008285 | involved_in negative regulation of cell population proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0009653 | involved_in anatomical structure morphogenesis | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0010614 | involved_in negative regulation of cardiac muscle hypertrophy | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0010631 | involved_in epithelial cell migration | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0010666 | involved_in positive regulation of cardiac muscle cell apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010718 | involved_in positive regulation of epithelial to mesenchymal transition | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0010862 | involved_in positive regulation of pathway-restricted SMAD protein phosphorylation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0014033 | involved_in neural crest cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0017015 | involved_in regulation of transforming growth factor beta receptor signaling pathway | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0030154 | involved_in cell differentiation | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0030308 | involved_in negative regulation of cell growth | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0030325 | involved_in adrenal gland development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0030509 | involved_in BMP signaling pathway | 3 | IBA,IDA,ISS | Homo_sapiens(9606) | Process |
| GO:0030511 | involved_in positive regulation of transforming growth factor beta receptor signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0030513 | involved_in positive regulation of BMP signaling pathway | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0031005 | enables filamin binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0032444 | part_of activin responsive factor complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0032525 | involved_in somite rostral/caudal axis specification | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0032909 | involved_in regulation of transforming growth factor beta2 production | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032924 | involved_in activin receptor signaling pathway | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0033686 | involved_in positive regulation of luteinizing hormone secretion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0035556 | involved_in intracellular signal transduction | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0036302 | involved_in atrioventricular canal development | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0042118 | involved_in endothelial cell activation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0042177 | involved_in negative regulation of protein catabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0042733 | involved_in embryonic digit morphogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0042802 | enables identical protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0042803 | enables protein homodimerization activity | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0043199 | enables sulfate binding | 1 | IMP | Homo_sapiens(9606) | Function |
| GO:0043565 | contributes_to sequence-specific DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0045892 | involved_in negative regulation of DNA-templated transcription | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045893 | involved_in positive regulation of DNA-templated transcription | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 2 | IDA,ISS | Homo_sapiens(9606) | Process |
| GO:0046872 | enables metal ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0046881 | involved_in positive regulation of follicle-stimulating hormone secretion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0048382 | involved_in mesendoderm development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0048589 | involved_in developmental growth | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0048663 | involved_in neuron fate commitment | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0048733 | involved_in sebaceous gland development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0048859 | involved_in formation of anatomical boundary | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051098 | involved_in regulation of binding | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051571 | involved_in positive regulation of histone H3-K4 methylation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0051797 | involved_in regulation of hair follicle development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060065 | involved_in uterus development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060391 | involved_in positive regulation of SMAD protein signal transduction | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0060395 | involved_in SMAD protein signal transduction | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0060412 | involved_in ventricular septum morphogenesis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0060956 | involved_in endocardial cell differentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0061040 | involved_in female gonad morphogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0061629 | enables RNA polymerase II-specific DNA-binding transcription factor binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0062009 | involved_in secondary palate development | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0070102 | involved_in interleukin-6-mediated signaling pathway | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0070371 | involved_in ERK1 and ERK2 cascade | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0070373 | involved_in negative regulation of ERK1 and ERK2 cascade | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0070411 | enables I-SMAD binding | 2 | IBA,IPI | Homo_sapiens(9606) | Function |
| GO:0070412 | enables R-SMAD binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0071141 | part_of SMAD protein complex | 2 | IDA,IMP | Homo_sapiens(9606) | Component |
| GO:0071144 | part_of heteromeric SMAD protein complex | 2 | IBA,IPI | Homo_sapiens(9606) | Component |
| GO:0071333 | involved_in cellular response to glucose stimulus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071559 | involved_in response to transforming growth factor beta | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071773 | involved_in cellular response to BMP stimulus | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0072133 | involved_in metanephric mesenchyme morphogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0072134 | involved_in nephrogenic mesenchyme morphogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0072520 | involved_in seminiferous tubule development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1901522 | involved_in positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:1902895 | involved_in positive regulation of miRNA transcription | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1905305 | involved_in negative regulation of cardiac myofibril assembly | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:2000617 | involved_in positive regulation of histone H3-K9 acetylation | 1 | ISS | Homo_sapiens(9606) | Process |