RBP Type
Canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
E3 ligase
Localizations
Stress granucle
BulkPerturb-seq
Ensembl ID ENSG00000141569 Gene ID
201292 Accession
27316
Symbol
TRIM65
Alias
4732463G12Rik
Full Name
tripartite motif containing 65
Status
Confidence
Length
16617 bases
Strand
Minus strand
Position
17 : 75880335 - 75896951
RNA binding domain
SPRY
Summary
Predicted to enable zinc ion binding activity. Involved in positive regulation of autophagy. Located in cytosol and nucleoplasm. [provided by Alliance of Genome Resources, Apr 2022]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000269383
SPRY
PF00622 2.4e-06
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
33373584 Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Kazuki Kato
2021-02-04
Molecular cell
Name
Transcript ID
bp
Protein
Translation ID
TRIM65-205
ENST00000591668
644
137aa
ENSP00000465034
TRIM65-206
ENST00000592642
396
90aa
ENSP00000466353
TRIM65-201
ENST00000269383
3384
517aa
ENSP00000269383
TRIM65-207
ENST00000648382
2049
No protein
-
TRIM65-204
ENST00000543309
865
288aa
ENSP00000441480
TRIM65-202
ENST00000540128
886
244aa
ENSP00000444607
TRIM65-203
ENST00000540812
385
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
37125 Platelet Function Tests 4.6000000E-007 - 37506 Platelet Function Tests 1.8600000E-006 - 39462 Platelet Function Tests 6.1800000E-007 - 39486 Platelet Function Tests 6.5200000E-006 - 72720 Brain Mapping 4E-15 21681796
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
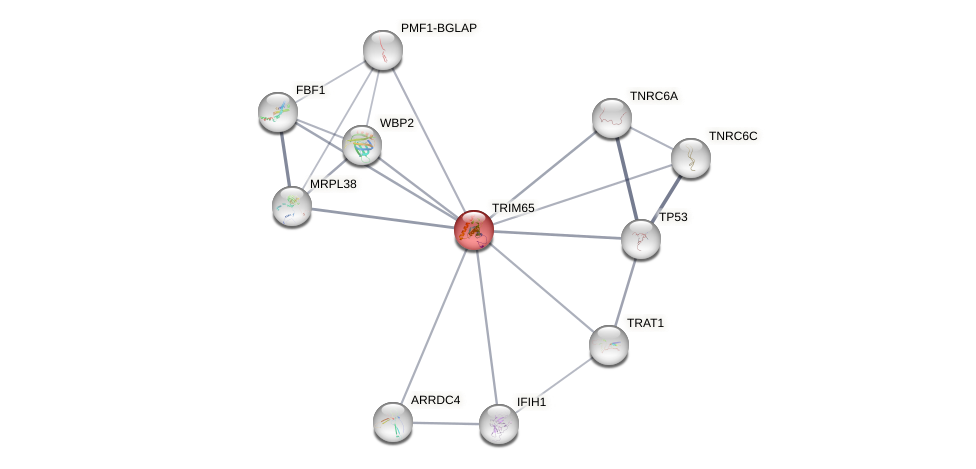
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0001934 involved_in positive regulation of protein phosphorylation 1 IEA Homo_sapiens(9606) Process GO:0005654 located_in nucleoplasm 1 IDA Homo_sapiens(9606) Component GO:0005737 is_active_in cytoplasm 1 IDA Homo_sapiens(9606) Component GO:0005829 located_in cytosol 1 IDA Homo_sapiens(9606) Component GO:0008270 enables zinc ion binding 1 IEA Homo_sapiens(9606) Function GO:0010508 involved_in positive regulation of autophagy 2 IBA,IMP Homo_sapiens(9606) Process GO:0032461 involved_in positive regulation of protein oligomerization 1 IEA Homo_sapiens(9606) Process GO:0032727 involved_in positive regulation of interferon-alpha production 1 IEA Homo_sapiens(9606) Process GO:0032728 involved_in positive regulation of interferon-beta production 1 IDA Homo_sapiens(9606) Process GO:0050728 involved_in negative regulation of inflammatory response 1 IDA Homo_sapiens(9606) Process GO:0060337 involved_in type I interferon-mediated signaling pathway 1 IEA Homo_sapiens(9606) Process GO:0061630 enables ubiquitin protein ligase activity 1 IDA Homo_sapiens(9606) Function GO:0070534 involved_in protein K63-linked ubiquitination 1 IDA Homo_sapiens(9606) Process GO:0070936 involved_in protein K48-linked ubiquitination 1 IDA Homo_sapiens(9606) Process GO:0140374 involved_in antiviral innate immune response 1 IEA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.