Welcome to
RBP World!
Functions and Diseases
| RBP Type | Non-canonical_RBPs |
| Diseases | CancerCHIKVFlavivirusesHIVSARS-COV-2SINVDengueZika |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | Metabolic enzyme |
| Localizations | Stress granucle |
| BulkPerturb-seq | N.A. |
Description
| Ensembl ID | ENSG00000132341 | Gene ID | 5901 | Accession | 9846 |
| Symbol | RAN | Alias | TC4;Gsp1;ARA24 | Full Name | RAN, member RAS oncogene family |
| Status | Confidence | Length | 5642 bases | Strand | Plus strand |
| Position | 12 : 130872037 - 130877678 | RNA binding domain | N.A. | ||
| Summary | RAN (ras-related nuclear protein) is a small GTP binding protein belonging to the RAS superfamily that is essential for the translocation of RNA and proteins through the nuclear pore complex. The RAN protein is also involved in control of DNA synthesis and cell cycle progression. Nuclear localization of RAN requires the presence of regulator of chromosome condensation 1 (RCC1). Mutations in RAN disrupt DNA synthesis. Because of its many functions, it is likely that RAN interacts with several other proteins. RAN regulates formation and organization of the microtubule network independently of its role in the nucleus-cytosol exchange of macromolecules. RAN could be a key signaling molecule regulating microtubule polymerization during mitosis. RCC1 generates a high local concentration of RAN-GTP around chromatin which, in turn, induces the local nucleation of microtubules. RAN is an androgen receptor (AR) coactivator that binds differentially with different lengths of polyglutamine within the androgen receptor. Polyglutamine repeat expansion in the AR is linked to Kennedy's disease (X-linked spinal and bulbar muscular atrophy). RAN coactivation of the AR diminishes with polyglutamine expansion within the AR, and this weak coactivation may lead to partial androgen insensitivity during the development of Kennedy's disease. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| RAN-203 | ENST00000448750 | 2455 | 234aa | ENSP00000396127 |
| RAN-207 | ENST00000536606 | 572 | 103aa | ENSP00000437950 |
| RAN-208 | ENST00000537745 | 1065 | No protein | - |
| RAN-210 | ENST00000541630 | 1475 | 128aa | ENSP00000441210 |
| RAN-212 | ENST00000543796 | 2474 | 216aa | ENSP00000446215 |
| RAN-205 | ENST00000477395 | 762 | No protein | - |
| RAN-202 | ENST00000392369 | 1530 | 216aa | ENSP00000376176 |
| RAN-206 | ENST00000535090 | 657 | 198aa | ENSP00000444042 |
| RAN-211 | ENST00000541679 | 572 | 53aa | ENSP00000483687 |
| RAN-204 | ENST00000464211 | 562 | No protein | - |
| RAN-201 | ENST00000392367 | 780 | 233aa | ENSP00000376174 |
| RAN-209 | ENST00000539498 | 1659 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 30237 | Calcium | 5.0170000E-005 | - |
| 86704 | Panic Disorder | 7E-6 | 19165232 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
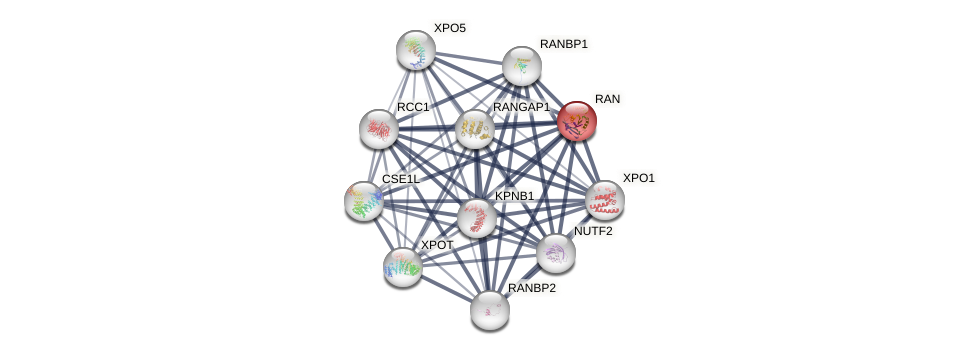
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000132341 | ||||||||
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | nomascus_leucogenys | ENSNLEP00000033817 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | pan_paniscus | ENSPPAP00000015741 | 92.7039 | 92.7039 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | ornithorhynchus_anatinus | ENSOANP00000010293 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | sarcophilus_harrisii | ENSSHAP00000036564 | 95.1327 | 94.2478 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 98.6111 | 97.6852 | sarcophilus_harrisii | ENSSHAP00000021258 | 98.6111 | 97.6852 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 62.963 | 60.1852 | notamacropus_eugenii | ENSMEUP00000004172 | 90.0662 | 86.0927 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 87.963 | 87.5 | choloepus_hoffmanni | ENSCHOP00000010316 | 99.4764 | 98.9529 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | dasypus_novemcinctus | ENSDNOP00000007138 | 99.0826 | 99.0826 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 61.5741 | 61.5741 | echinops_telfairi | ENSETEP00000009634 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 43.0556 | 38.4259 | echinops_telfairi | ENSETEP00000007392 | 85.3211 | 76.1468 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | tursiops_truncatus | ENSTTRP00000012079 | 93.5065 | 93.5065 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | vulpes_vulpes | ENSVVUP00000035721 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 96.7593 | 96.7593 | capra_hircus | ENSCHIP00000028737 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | loxodonta_africana | ENSLAFP00000014817 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 59.2593 | 59.2593 | ailuropoda_melanoleuca | ENSAMEP00000038509 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | equus_caballus | ENSECAP00000031233 | 66.8731 | 66.8731 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | balaenoptera_musculus | ENSBMSP00010028991 | 66.6667 | 66.6667 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | felis_catus | ENSFCAP00000019194 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 96.7593 | 93.9815 | monodelphis_domestica | ENSMODP00000008542 | 97.2093 | 94.4186 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | monodon_monoceros | ENSMMNP00015020999 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | jaculus_jaculus | ENSJJAP00000007631 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | physeter_catodon | ENSPCTP00005024907 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | mus_caroli | MGP_CAROLIEiJ_P0071016 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | mus_musculus | ENSMUSP00000106975 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 57.4074 | 56.9444 | aotus_nancymaae | ENSANAP00000003754 | 96.875 | 96.0938 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 95.3704 | 91.2037 | aotus_nancymaae | ENSANAP00000037675 | 94.9309 | 90.7834 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | saimiri_boliviensis_boliviensis | ENSSBOP00000015309 | 92.7039 | 92.7039 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 67.5926 | 67.5926 | chlorocebus_sabaeus | ENSCSAP00000016123 | 96.0526 | 96.0526 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 50.463 | 50.463 | tupaia_belangeri | ENSTBEP00000000995 | 64.497 | 64.497 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | rattus_norvegicus | ENSRNOP00000001247 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | bos_mutus | ENSBMUP00000000488 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | mus_spicilegus | ENSMSIP00000005823 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | vombatus_ursinus | ENSVURP00010004850 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | rhinolophus_ferrumequinum | ENSRFEP00010027260 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.4444 | 92.5926 | neovison_vison | ENSNVIP00000020998 | 98.0769 | 96.1538 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | canis_lupus_dingo | ENSCAFP00020014073 | 96.4286 | 96.4286 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | phascolarctos_cinereus | ENSPCIP00000029058 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 95.3704 | 92.5926 | phascolarctos_cinereus | ENSPCIP00000025970 | 95.3704 | 92.5926 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | nannospalax_galili | ENSNGAP00000009532 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | bos_grunniens | ENSBGRP00000014545 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | cervus_hanglu_yarkandensis | ENSCHYP00000005856 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | cervus_hanglu_yarkandensis | ENSCHYP00000029759 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 94.9074 | ictidomys_tridecemlineatus | ENSSTOP00000017027 | 97.619 | 97.619 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | otolemur_garnettii | ENSOGAP00000019924 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | moschus_moschiferus | ENSMMSP00000015596 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | rhinopithecus_roxellana | ENSRROP00000044160 | 92.7039 | 92.7039 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 100 | 100 | mus_pahari | MGP_PahariEiJ_P0059216 | 100 | 100 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 91.6667 | 87.037 | caenorhabditis_elegans | K01G5.4.1 | 92.093 | 87.4419 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 75 | 60.1852 | drosophila_melanogaster | FBpp0075369 | 74.6544 | 59.9078 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 67.5926 | 65.2778 | ciona_intestinalis | ENSCINP00000008166 | 90.6832 | 87.5776 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 78.2407 | 76.8519 | petromyzon_marinus | ENSPMAP00000002493 | 96.0227 | 94.3182 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 95.3704 | 91.2037 | eptatretus_burgeri | ENSEBUP00000017437 | 75.4579 | 72.1612 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 40.2778 | 38.4259 | callorhinchus_milii | ENSCMIP00000020451 | 71.9008 | 68.595 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 65.7407 | 64.3519 | latimeria_chalumnae | ENSLACP00000021390 | 96.5986 | 94.5578 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 98.6111 | 96.7593 | lepisosteus_oculatus | ENSLOCP00000007531 | 96.8182 | 95 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 96.7593 | 94.4444 | clupea_harengus | ENSCHAP00000005823 | 85.6557 | 83.6066 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 96.7593 | 94.4444 | clupea_harengus | ENSCHAP00000002546 | 88.5593 | 86.4407 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 96.2963 | 94.4444 | danio_rerio | ENSDARP00000108629 | 96.7442 | 94.8837 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 97.2222 | 94.9074 | carassius_auratus | ENSCARP00000034230 | 73.9437 | 72.1831 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 96.7593 | 94.4444 | carassius_auratus | ENSCARP00000079950 | 89.3162 | 87.1795 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 96.7593 | 93.9815 | astyanax_mexicanus | ENSAMXP00000053248 | 97.2093 | 94.4186 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 95.3704 | 92.5926 | ictalurus_punctatus | ENSIPUP00000023414 | 89.1775 | 86.5801 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 91.2037 | 87.963 | esox_lucius | ENSELUP00000074648 | 90.367 | 87.156 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 89.8148 | 86.5741 | esox_lucius | ENSELUP00000026440 | 88.9908 | 85.7798 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.5185 | 90.7407 | electrophorus_electricus | ENSEEEP00000031951 | 93.0876 | 90.3226 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 92.1296 | 89.3519 | oncorhynchus_kisutch | ENSOKIP00005033665 | 92.5581 | 89.7674 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 98.1481 | 95.3704 | oncorhynchus_kisutch | ENSOKIP00005053554 | 98.1481 | 95.3704 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 98.1481 | 95.3704 | oncorhynchus_kisutch | ENSOKIP00005025125 | 95.0673 | 92.3767 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.0556 | 88.8889 | oncorhynchus_kisutch | ENSOKIP00005033501 | 83.0579 | 79.3388 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.4444 | 92.1296 | oncorhynchus_mykiss | ENSOMYP00000040548 | 89.4737 | 87.2807 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | oncorhynchus_mykiss | ENSOMYP00000072115 | 91.5179 | 87.9464 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 98.1481 | 95.3704 | oncorhynchus_mykiss | ENSOMYP00000116809 | 85.83 | 83.4008 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 90.7407 | 87.963 | oncorhynchus_mykiss | ENSOMYP00000041802 | 71.2727 | 69.0909 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 95.8333 | 90.7407 | salmo_salar | ENSSSAP00000059417 | 86.25 | 81.6667 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.6667 | salmo_salar | ENSSSAP00000164345 | 84.0164 | 81.1475 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 98.1481 | 95.3704 | salmo_salar | ENSSSAP00000059421 | 92.9825 | 90.3509 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.4444 | 91.6667 | salmo_salar | ENSSSAP00000007579 | 92.3077 | 89.5928 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.4444 | 90.7407 | salmo_trutta | ENSSTUP00000069436 | 94.4444 | 90.7407 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.4444 | 91.6667 | salmo_trutta | ENSSTUP00000080058 | 87.1795 | 84.6154 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 90.7407 | 87.963 | salmo_trutta | ENSSTUP00000100003 | 68.2927 | 66.2021 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 98.1481 | 95.3704 | salmo_trutta | ENSSTUP00000041469 | 94.6429 | 91.9643 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 92.5926 | 89.3519 | gadus_morhua | ENSGMOP00000023762 | 87.3362 | 84.2795 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | fundulus_heteroclitus | ENSFHEP00000014016 | 93.6073 | 89.9543 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 90.2778 | poecilia_reticulata | ENSPREP00000025717 | 94.0367 | 89.4495 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | xiphophorus_maculatus | ENSXMAP00000009949 | 93.6073 | 89.9543 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | oryzias_latipes | ENSORLP00000004290 | 93.6073 | 89.9543 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 92.5926 | 88.8889 | cyclopterus_lumpus | ENSCLMP00005020958 | 88.8889 | 85.3333 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | oreochromis_niloticus | ENSONIP00000031362 | 78.8462 | 75.7692 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 86.5741 | 81.0185 | haplochromis_burtoni | ENSHBUP00000027609 | 76.9547 | 72.0165 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.5185 | 88.8889 | astatotilapia_calliptera | ENSACLP00000020685 | 87.8261 | 83.4783 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.9815 | 90.7407 | sparus_aurata | ENSSAUP00010022102 | 87.5 | 84.4828 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.9815 | 90.2778 | lates_calcarifer | ENSLCAP00010024131 | 83.8843 | 80.5785 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 98.6111 | 98.1481 | xenopus_tropicalis | ENSXETP00000049065 | 98.6111 | 98.1481 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | chrysemys_picta_bellii | ENSCPBP00000035610 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 87.5 | 86.1111 | crocodylus_porosus | ENSCPRP00005027001 | 85.5204 | 84.1629 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.9815 | 93.0556 | laticauda_laticaudata | ENSLLTP00000005319 | 99.5098 | 98.5294 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | notechis_scutatus | ENSNSUP00000019191 | 99.0783 | 98.1567 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | pseudonaja_textilis | ENSPTXP00000026646 | 99.0783 | 98.1567 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000013416 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | gallus_gallus | ENSGALP00010034211 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 96.7593 | 95.3704 | serinus_canaria | ENSSCAP00000015576 | 92.8889 | 91.5556 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | parus_major | ENSPMJP00000001214 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | anolis_carolinensis | ENSACAP00000007043 | 84.3137 | 83.5294 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | neolamprologus_brichardi | ENSNBRP00000005922 | 95.3488 | 91.6279 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | naja_naja | ENSNNAP00000024905 | 99.0783 | 98.1567 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | podarcis_muralis | ENSPMRP00000027024 | 99.0783 | 98.1567 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | geospiza_fortis | ENSGFOP00000005402 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | taeniopygia_guttata | ENSTGUP00000004833 | 54.5685 | 54.0609 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 97.2222 | 96.2963 | erpetoichthys_calabaricus | ENSECRP00000015504 | 97.6744 | 96.7442 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | kryptolebias_marmoratus | ENSKMAP00000015434 | 93.6073 | 89.9543 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.0741 | 98.1481 | salvator_merianae | ENSSMRP00000008420 | 54.0404 | 53.5354 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.5185 | 89.3519 | oryzias_javanicus | ENSOJAP00000013288 | 91.4027 | 87.3303 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | amphilophus_citrinellus | ENSACIP00000008572 | 82.3293 | 79.1165 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 92.5926 | 89.3519 | dicentrarchus_labrax | ENSDLAP00005033650 | 90.9091 | 87.7273 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.9815 | 91.2037 | amphiprion_percula | ENSAPEP00000016410 | 92.6941 | 89.9543 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.4444 | 90.7407 | oryzias_sinensis | ENSOSIP00000029081 | 91.0714 | 87.5 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | pelodiscus_sinensis | ENSPSIP00000002413 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | pelodiscus_sinensis | ENSPSIP00000012797 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | gopherus_evgoodei | ENSGEVP00005029356 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | chelonoidis_abingdonii | ENSCABP00000021729 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.4444 | 91.2037 | seriola_dumerili | ENSSDUP00000020401 | 94.8837 | 91.6279 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.4444 | 89.8148 | cottoperca_gobio | ENSCGOP00000013622 | 91.8919 | 87.3874 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 95.3704 | 93.5185 | struthio_camelus_australis | ENSSCUP00000005294 | 89.9563 | 88.2096 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | anser_brachyrhynchus | ENSABRP00000020128 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.4444 | 91.2037 | gasterosteus_aculeatus | ENSGACP00000023469 | 94.4444 | 91.2037 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.0556 | 88.4259 | cynoglossus_semilaevis | ENSCSEP00000010989 | 77.6062 | 73.7452 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | poecilia_formosa | ENSPFOP00000009579 | 89.5197 | 86.0262 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.9815 | 90.2778 | myripristis_murdjan | ENSMMDP00005018277 | 81.5261 | 78.3133 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.4444 | 90.7407 | sander_lucioperca | ENSSLUP00000048320 | 85.7143 | 82.3529 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | strigops_habroptila | ENSSHBP00005018753 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.9815 | 91.2037 | amphiprion_ocellaris | ENSAOCP00000024318 | 92.6941 | 89.9543 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | terrapene_carolina_triunguis | ENSTMTP00000028978 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 90.7407 | 87.963 | hucho_hucho | ENSHHUP00000019483 | 76.5625 | 74.2188 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 92.5926 | 88.8889 | hucho_hucho | ENSHHUP00000065899 | 82.9875 | 79.6681 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 92.1296 | 87.5 | hucho_hucho | ENSHHUP00000036805 | 79.6 | 75.6 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 90.7407 | hucho_hucho | ENSHHUP00000018425 | 82 | 78.4 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.5185 | 87.963 | betta_splendens | ENSBSLP00000016341 | 95.283 | 89.6226 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 96.7593 | 93.9815 | pygocentrus_nattereri | ENSPNAP00000019424 | 96.7593 | 93.9815 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 92.1296 | 88.4259 | anabas_testudineus | ENSATEP00000041732 | 87.2807 | 83.7719 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 89.3519 | 81.9444 | saccharomyces_cerevisiae | YOR185C | 87.7273 | 80.4545 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 88.8889 | 81.9444 | saccharomyces_cerevisiae | YLR293C | 87.6712 | 80.8219 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.4444 | 91.2037 | seriola_lalandi_dorsalis | ENSSLDP00000001645 | 94.8837 | 91.6279 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.9815 | 89.3519 | labrus_bergylta | ENSLBEP00000012035 | 94.4186 | 89.7674 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.9815 | 89.3519 | labrus_bergylta | ENSLBEP00000020980 | 94.4186 | 89.7674 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | pundamilia_nyererei | ENSPNYP00000022194 | 95.3488 | 91.6279 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | mastacembelus_armatus | ENSMAMP00000061586 | 94.9074 | 91.2037 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | stegastes_partitus | ENSSPAP00000020865 | 95.3488 | 91.6279 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 98.1481 | 95.3704 | oncorhynchus_tshawytscha | ENSOTSP00005044846 | 83.7945 | 81.4229 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 97.6852 | 94.4444 | oncorhynchus_tshawytscha | ENSOTSP00005025340 | 94.6188 | 91.4798 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 92.5926 | 88.4259 | oncorhynchus_tshawytscha | ENSOTSP00005034533 | 77.5194 | 74.031 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.9815 | 90.2778 | oncorhynchus_tshawytscha | ENSOTSP00005015146 | 80.8765 | 77.6892 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.4444 | 91.2037 | acanthochromis_polyacanthus | ENSAPOP00000024679 | 94.8837 | 91.6279 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | aquila_chrysaetos_chrysaetos | ENSACCP00020002204 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 92.5926 | 87.5 | nothobranchius_furzeri | ENSNFUP00015006832 | 90.9091 | 85.9091 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | cyprinodon_variegatus | ENSCVAP00000017476 | 93.6073 | 89.9543 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 96.7593 | 94.4444 | denticeps_clupeoides | ENSDCDP00000018053 | 90.0862 | 87.931 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.5185 | 89.8148 | larimichthys_crocea | ENSLCRP00005017121 | 87.8261 | 84.3478 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.9815 | 92.5926 | takifugu_rubripes | ENSTRUP00000035114 | 90.625 | 89.2857 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | ficedula_albicollis | ENSFALP00000030206 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.0556 | 89.3519 | hippocampus_comes | ENSHCOP00000007683 | 93.0556 | 89.3519 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 96.7593 | 94.4444 | cyprinus_carpio_carpio | ENSCCRP00000038653 | 83.6 | 81.6 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 97.2222 | 94.4444 | cyprinus_carpio_carpio | ENSCCRP00000038657 | 90.9091 | 88.3117 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 99.537 | 98.6111 | coturnix_japonica | ENSCJPP00005024964 | 99.537 | 98.6111 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | poecilia_latipinna | ENSPLAP00000000760 | 93.6073 | 89.9543 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.5185 | 89.8148 | sinocyclocheilus_grahami | ENSSGRP00000033662 | 91.4027 | 87.7828 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.9815 | 91.2037 | sinocyclocheilus_grahami | ENSSGRP00000000281 | 93.9815 | 91.2037 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | maylandia_zebra | ENSMZEP00005004282 | 95.3488 | 91.6279 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.5185 | 91.2037 | tetraodon_nigroviridis | ENSTNIP00000016119 | 93.5185 | 91.2037 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 97.6852 | 96.2963 | paramormyrops_kingsleyae | ENSPKIP00000036577 | 98.1395 | 96.7442 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 97.2222 | 96.2963 | paramormyrops_kingsleyae | ENSPKIP00000000474 | 97.6744 | 96.7442 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 97.6852 | 96.2963 | scleropages_formosus | ENSSFOP00015001367 | 90.9483 | 89.6552 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 97.6852 | 95.8333 | scleropages_formosus | ENSSFOP00015011049 | 95.9091 | 94.0909 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 96.2963 | 94.9074 | leptobrachium_leishanense | ENSLLEP00000027885 | 96.2963 | 94.9074 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 90.7407 | 88.4259 | leptobrachium_leishanense | ENSLLEP00000011586 | 96.0784 | 93.6274 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 93.5185 | 88.8889 | scophthalmus_maximus | ENSSMAP00000047691 | 92.2374 | 87.6712 |
| ENSG00000132341 | homo_sapiens | ENSP00000446215 | 94.9074 | 91.2037 | oryzias_melastigma | ENSOMEP00000034237 | 93.6073 | 89.9543 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000054 | involved_in ribosomal subunit export from nucleus | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0000055 | involved_in ribosomal large subunit export from nucleus | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0000056 | involved_in ribosomal small subunit export from nucleus | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0000070 | involved_in mitotic sister chromatid segregation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0000278 | involved_in mitotic cell cycle | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0000287 | enables magnesium ion binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0001673 | located_in male germ cell nucleus | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0002177 | located_in manchette | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0003682 | enables chromatin binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0003924 | enables GTPase activity | 3 | IBA,IDA,TAS | Homo_sapiens(9606) | Function |
| GO:0003925 | enables G protein activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005049 | contributes_to nuclear export signal receptor activity | 1 | IC | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005525 | enables GTP binding | 2 | IDA,IMP | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 3 | HDA,IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005635 | located_in nuclear envelope | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005643 | part_of nuclear pore | 1 | NAS | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005730 | located_in nucleolus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005814 | located_in centriole | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0006259 | involved_in DNA metabolic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006606 | involved_in protein import into nucleus | 3 | IBA,IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0006611 | involved_in protein export from nucleus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0007052 | involved_in mitotic spindle organization | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007286 | involved_in spermatid development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0016020 | located_in membrane | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0016032 | involved_in viral process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0019003 | enables GDP binding | 2 | IDA,IMP | Homo_sapiens(9606) | Function |
| GO:0019904 | enables protein domain specific binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0021766 | involved_in hippocampus development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0030036 | involved_in actin cytoskeleton organization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0030496 | located_in midbody | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0032092 | involved_in positive regulation of protein binding | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032991 | part_of protein-containing complex | 2 | IDA,IMP | Homo_sapiens(9606) | Component |
| GO:0035281 | involved_in pre-miRNA export from nucleus | 1 | IC | Homo_sapiens(9606) | Process |
| GO:0036126 | located_in sperm flagellum | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0042307 | involved_in positive regulation of protein import into nucleus | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0042470 | located_in melanosome | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0042565 | part_of RNA nuclear export complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0045296 | enables cadherin binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0045505 | enables dynein intermediate chain binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0046039 | involved_in GTP metabolic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0046982 | enables protein heterodimerization activity | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0051301 | involved_in cell division | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0055037 | located_in recycling endosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0061015 | involved_in snRNA import into nucleus | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0061676 | enables importin-alpha family protein binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0070062 | located_in extracellular exosome | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0070883 | contributes_to pre-miRNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0071389 | involved_in cellular response to mineralocorticoid stimulus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0090543 | NOT located_in Flemming body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:1902570 | involved_in protein localization to nucleolus | 1 | IMP | Homo_sapiens(9606) | Process |