RBP Type
Canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
E3 ligase
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000132256 Gene ID
85363 Accession
16276
Symbol
TRIM5
Alias
RNF88;TRIM5alpha
Full Name
tripartite motif containing 5
Status
Confidence
Length
275425 bases
Strand
Minus strand
Position
11 : 5663195 - 5938619
RNA binding domain
SPRY
Summary
The protein encoded by this gene is a member of the tripartite motif (TRIM) family. The TRIM motif includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. The protein forms homo-oligomers via the coilel-coil region and localizes to cytoplasmic bodies. It appears to function as a E3 ubiquitin-ligase and ubiqutinates itself to regulate its subcellular localization. It may play a role in retroviral restriction. Multiple alternatively spliced transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Dec 2009]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000369373
SPRY
PF00622 1.4e-09
1
ENSP00000507420
SPRY
PF00622 1.4e-09
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
TRIM5-202
ENST00000380034
3364
493aa
ENSP00000369373
TRIM5-201
ENST00000380027
2290
326aa
ENSP00000369366
TRIM5-206
ENST00000433961
2759
271aa
ENSP00000393052
TRIM5-214
ENST00000684655
2862
493aa
ENSP00000507420
TRIM5-203
ENST00000396847
3170
347aa
ENSP00000380058
TRIM5-212
ENST00000682968
2669
265aa
ENSP00000506742
TRIM5-207
ENST00000438025
767
187aa
ENSP00000398196
TRIM5-209
ENST00000483835
918
No protein
-
TRIM5-211
ENST00000492086
662
No protein
-
TRIM5-213
ENST00000684417
1317
No protein
-
TRIM5-208
ENST00000465634
602
No protein
-
TRIM5-210
ENST00000487241
756
No protein
-
TRIM5-204
ENST00000412903
1054
209aa
ENSP00000388031
TRIM5-205
ENST00000419850
607
72aa
ENSP00000388150
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
18319 Fetal Hemoglobin 8.839e-005 - 18452 Hemoglobins 1.399e-005 - 61117 Immunoglobulins 2E-6 23382691 62914 Immunoglobulins 3E-6 23382691 77193 Immunoglobulins 1E-6 23382691
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
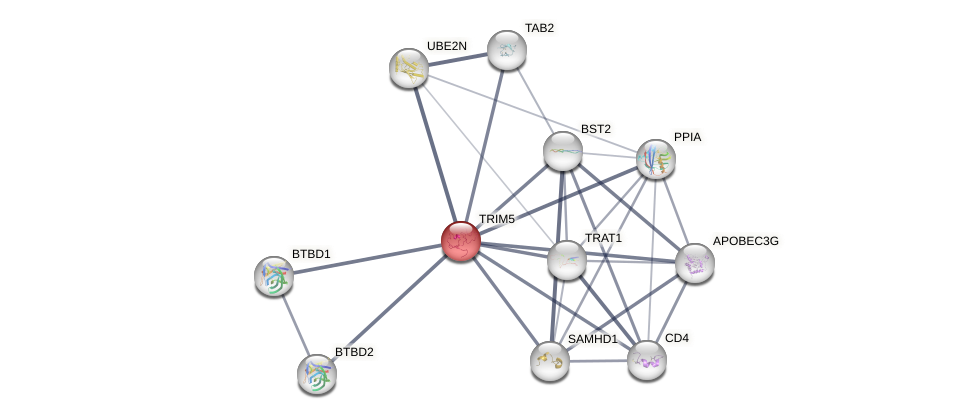
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000209 involved_in protein polyubiquitination 1 IBA Homo_sapiens(9606) Process GO:0000932 located_in P-body 1 IDA Homo_sapiens(9606) Component GO:0002218 involved_in activation of innate immune response 1 IDA Homo_sapiens(9606) Process GO:0003713 enables transcription coactivator activity 1 IDA Homo_sapiens(9606) Function GO:0004842 enables ubiquitin-protein transferase activity 1 IDA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005654 is_active_in nucleoplasm 1 IBA Homo_sapiens(9606) Component GO:0005737 is_active_in cytoplasm 2 IBA,IDA Homo_sapiens(9606) Component GO:0005829 is_active_in cytosol 2 IBA,TAS Homo_sapiens(9606) Component GO:0006914 involved_in autophagy 1 IDA Homo_sapiens(9606) Process GO:0008270 enables zinc ion binding 1 IEA Homo_sapiens(9606) Function GO:0010508 involved_in positive regulation of autophagy 1 IBA Homo_sapiens(9606) Process GO:0019901 enables protein kinase binding 2 IBA,IPI Homo_sapiens(9606) Function GO:0030674 enables protein-macromolecule adaptor activity 1 IPI Homo_sapiens(9606) Function GO:0031664 involved_in regulation of lipopolysaccharide-mediated signaling pathway 1 IMP Homo_sapiens(9606) Process GO:0032880 involved_in regulation of protein localization 2 IBA,IMP Homo_sapiens(9606) Process GO:0038187 enables pattern recognition receptor activity 1 IDA Homo_sapiens(9606) Function GO:0042802 enables identical protein binding 1 IPI Homo_sapiens(9606) Function GO:0042803 enables protein homodimerization activity 2 IBA,IPI Homo_sapiens(9606) Function GO:0043123 involved_in positive regulation of I-kappaB kinase/NF-kappaB signaling 3 IBA,IDA,IMP Homo_sapiens(9606) Process GO:0043410 involved_in positive regulation of MAPK cascade 1 IMP Homo_sapiens(9606) Process GO:0044790 involved_in suppression of viral release by host 1 IDA Homo_sapiens(9606) Process GO:0045087 involved_in innate immune response 2 IBA,IDA Homo_sapiens(9606) Process GO:0045893 involved_in positive regulation of DNA-templated transcription 1 IEA Homo_sapiens(9606) Process GO:0046596 involved_in regulation of viral entry into host cell 1 IBA Homo_sapiens(9606) Process GO:0046597 involved_in negative regulation of viral entry into host cell 1 IDA Homo_sapiens(9606) Process GO:0051091 involved_in positive regulation of DNA-binding transcription factor activity 1 IMP Homo_sapiens(9606) Process GO:0051092 involved_in positive regulation of NF-kappaB transcription factor activity 2 IBA,IMP Homo_sapiens(9606) Process GO:0051607 involved_in defense response to virus 1 TAS Homo_sapiens(9606) Process GO:0061630 enables ubiquitin protein ligase activity 1 IBA Homo_sapiens(9606) Function GO:0070534 involved_in protein K63-linked ubiquitination 1 IDA Homo_sapiens(9606) Process GO:1990462 colocalizes_with omegasome 1 IDA Homo_sapiens(9606) Component
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.