RBP Type
Canonical_RBPs
Diseases
N.A.
Drug
Main interacting RNAs
N.A.
Moonlighting functions
N.A.
Localizations
Nucleoli
BulkPerturb-seq
Ensembl ID ENSG00000129484 Gene ID
10038 Accession
272
Symbol
PARP2
Alias
ARTD2;ADPRT2;PARP-2;ADPRTL2;ADPRTL3;pADPRT-2
Full Name
poly(ADP-ribose) polymerase 2
Status
Confidence
Length
14290 bases
Strand
Plus strand
Position
14 : 20343615 - 20357904
RNA binding domain
PARP
Summary
This gene encodes poly(ADP-ribosyl)transferase-like 2 protein, which contains a catalytic domain and is capable of catalyzing a poly(ADP-ribosyl)ation reaction. This protein has a catalytic domain which is homologous to that of poly (ADP-ribosyl) transferase, but lacks an N-terminal DNA binding domain which activates the C-terminal catalytic domain of poly (ADP-ribosyl) transferase. The basic residues within the N-terminal region of this protein may bear potential DNA-binding properties, and may be involved in the nuclear and/or nucleolar targeting of the protein. Two alternatively spliced transcript variants encoding distinct isoforms have been found. [provided by RefSeq, Jul 2008]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000250416
PARP
PF00644 6.7e-70
1
ENSP00000392972
PARP
PF00644 8.8e-70
1
ENSP00000432283
PARP
PF00644 8.5e-56
1
ENSP00000445524
PARP
PF00644 4.6e-45
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
26091344 RNA Regulation by Poly(ADP-Ribose) Polymerases.
J Florian Bock
2015-06-18
Molecular cell
32656996 The multifaceted role of PARP1 in RNA biogenesis.
Rebekah Eleazer
2021-03-01
Wiley interdisciplinary reviews. RNA
31829559 Nonspecific Binding of RNA to PARP1 and PARP2 Does Not Lead to Catalytic Activation.
Y Meagan Nakamoto
2019-12-24
Biochemistry
33693821 An RNA tagging approach for system-wide RNA-binding proteome profiling and dynamics investigation upon transcription inhibition.
Zheng Zhang
2021-06-21
Nucleic acids research
24928857 PARP-2 and PARP-3 are selectively activated by 5' phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1.
Marie-France Langelier
2014-07-01
Nucleic acids research
37609662 The dynamic process of covalent and non-covalent PARylation in the maintenance of genome integrity: a focus on PARP inhibitors.
Adèle Beneyton
2023-09-01
NAR cancer
35349716 PARP inhibitors trap PARP2 and alter the mode of recruitment of PARP2 at DNA damage sites.
Xiaohui Lin
2022-04-22
Nucleic acids research
32441764 Repositioning PARP inhibitors for SARS-CoV-2 infection(COVID-19); a new multi-pronged therapy for acute respiratory distress syndrome?
Nicola Curtin
2020-08-01
British journal of pharmacology
32029452 PARPs and ADP-ribosylation in RNA biology: from RNA expression and processing to protein translation and proteostasis.
Dae-Seok Kim
2020-03-01
Genes & development
Name
Transcript ID
bp
Protein
Translation ID
PARP2-202
ENST00000429687
1827
570aa
ENSP00000392972
PARP2-201
ENST00000250416
1886
583aa
ENSP00000250416
PARP2-206
ENST00000529465
717
110aa
ENSP00000431867
PARP2-204
ENST00000527915
1980
531aa
ENSP00000432283
PARP2-205
ENST00000528465
551
No protein
-
PARP2-211
ENST00000555140
530
No protein
-
PARP2-203
ENST00000527384
429
No protein
-
PARP2-208
ENST00000532299
876
No protein
-
PARP2-210
ENST00000539930
635
177aa
ENSP00000445524
PARP2-207
ENST00000530598
842
No protein
-
PARP2-209
ENST00000534664
566
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
8428 Alcohol Drinking 1.38358566389269E-11 -
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
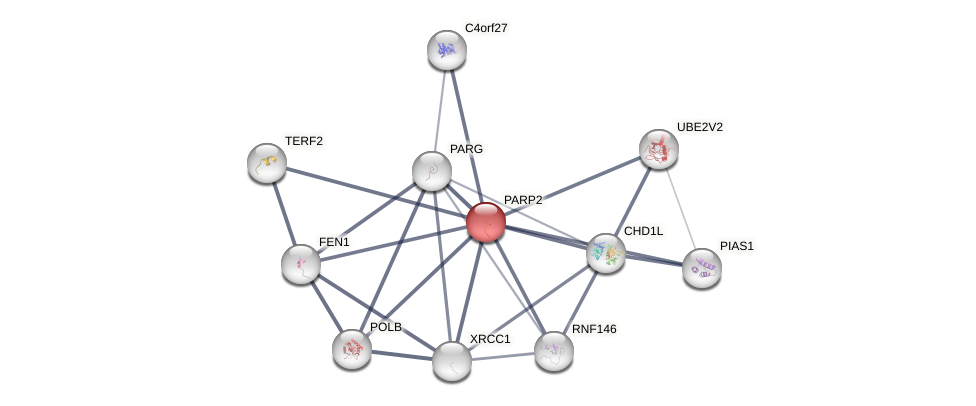
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0003682 enables chromatin binding 1 IDA Homo_sapiens(9606) Function GO:0003684 enables damaged DNA binding 1 IDA Homo_sapiens(9606) Function GO:0003950 enables NAD+ ADP-ribosyltransferase activity 2 IBA,IDA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 located_in nucleus 1 IDA Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 1 IDA Homo_sapiens(9606) Component GO:0005730 is_active_in nucleolus 2 IBA,IDA Homo_sapiens(9606) Component GO:0006281 involved_in DNA repair 1 IDA Homo_sapiens(9606) Process GO:0006284 involved_in base-excision repair 1 IEA Homo_sapiens(9606) Process GO:0006302 involved_in double-strand break repair 2 IBA,IDA Homo_sapiens(9606) Process GO:0006974 involved_in DNA damage response 1 IDA Homo_sapiens(9606) Process GO:0016779 enables nucleotidyltransferase activity 1 IEA Homo_sapiens(9606) Function GO:0030592 involved_in DNA ADP-ribosylation 1 IDA Homo_sapiens(9606) Process GO:0031491 enables nucleosome binding 1 IDA Homo_sapiens(9606) Function GO:0046697 involved_in decidualization 1 ISS Homo_sapiens(9606) Process GO:0061051 involved_in positive regulation of cell growth involved in cardiac muscle cell development 1 IEA Homo_sapiens(9606) Process GO:0070212 involved_in protein poly-ADP-ribosylation 2 IBA,IDA Homo_sapiens(9606) Process GO:0070213 involved_in protein auto-ADP-ribosylation 1 IDA Homo_sapiens(9606) Process GO:0072572 enables poly-ADP-D-ribose binding 1 IDA Homo_sapiens(9606) Function GO:0090734 is_active_in site of DNA damage 2 IDA Homo_sapiens(9606) Component GO:0097191 involved_in extrinsic apoptotic signaling pathway 1 IEA Homo_sapiens(9606) Process GO:0140294 enables NAD DNA ADP-ribosyltransferase activity 1 IDA Homo_sapiens(9606) Function GO:0140805 enables NAD+-protein-serine ADP-ribosyltransferase activity 1 IDA Homo_sapiens(9606) Function GO:0140806 enables NAD+- protein-aspartate ADP-ribosyltransferase activity 1 IDA Homo_sapiens(9606) Function GO:0140807 enables NAD+-protein-glutamate ADP-ribosyltransferase activity 1 IDA Homo_sapiens(9606) Function GO:0140861 involved_in DNA repair-dependent chromatin remodeling 1 IDA Homo_sapiens(9606) Process GO:0160004 enables poly-ADP-D-ribose modification-dependent protein binding 1 IDA Homo_sapiens(9606) Function GO:1901215 involved_in negative regulation of neuron death 1 IEA Homo_sapiens(9606) Process GO:1990404 enables NAD+-protein ADP-ribosyltransferase activity 2 IBA,IDA Homo_sapiens(9606) Function
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.