Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | Cancer |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | N.A. |
| Localizations | Stress granucle |
| BulkPerturb-seq | N.A. |
Description
| Ensembl ID | ENSG00000122870 | Gene ID | 80114 | Accession | 19351 |
| Symbol | BICC1 | Alias | BICC;CYSRD | Full Name | BicC family RNA binding protein 1 |
| Status | Confidence | Length | 318564 bases | Strand | Plus strand |
| Position | 10 : 58512872 - 58831435 | RNA binding domain | KH_1 | ||
| Summary | This gene encodes an RNA-binding protein that is active in regulating gene expression by modulating protein translation during embryonic development. Mouse studies identified the corresponding protein to be under strict control during cell differentiation and to be a maternally provided gene product. [provided by RefSeq, Apr 2009] | ||||
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 36531729 | Construction of a diagnostic signature and immune landscape of pulmonary arterial hypertension. | Mengjie Duo | 2022-01-01 | Frontiers in cardiovascular medicine |
| 29382913 | Human Skeletal Muscle Possesses an Epigenetic Memory of Hypertrophy. | A Robert Seaborne | 2018-01-30 | Scientific reports |
| 33258470 | Survival prediction in patients with colon adenocarcinoma via multi-omics data integration using a deep learning algorithm. | Jiudi Lv | 2020-12-01 | Bioscience reports |
| 36802443 | Deficiency of the minor spliceosome component U4atac snRNA secondarily results in ciliary defects in human and zebrafish. | Deepak Khatri | 2023-02-28 | Proceedings of the National Academy of Sciences of the United States of America |
| 30832715 | A comparison of DNA methylation in newborn blood samples from infants with and without orofacial clefts. | Zongli Xu | 2019-03-04 | Clinical epigenetics |
| 25715394 | Bicaudal C1 promotes pancreatic NEUROG3+ endocrine progenitor differentiation and ductal morphogenesis. | A Laurence Lemaire | 2015-03-01 | Development (Cambridge, England) |
| 36158189 | Bicaudal-C Post-transcriptional regulator of cell fates and functions. | E Megan Dowdle | 2022-01-01 | Frontiers in cell and developmental biology |
| 23884099 | microRNA biomarkers in cystic diseases. | Mi Yu Woo | 2013-07-01 | BMB reports |
| 29290488 | Crystal Structure of Bicc1 SAM Polymer and Mapping of Interactions between the Ciliopathy-Associated Proteins Bicc1, ANKS3, and ANKS6. | Benjamin Rothé | 2018-02-06 | Structure (London, England : 1993) |
| 25274489 | Transcriptome-wide landscape of pre-mRNA alternative splicing associated with metastatic colonization. | Zhi-xiang Lu | 2015-02-01 | Molecular cancer research : MCR |
| 37443111 | BICC1 drives pancreatic cancer progression by inducing VEGF-independent angiogenesis. | Chongbiao Huang | 2023-07-14 | Signal transduction and targeted therapy |
| 25888842 | Urine Fetuin-A is a biomarker of autosomal dominant polycystic kidney disease progression. | Nathalie Piazzon | 2015-03-30 | Journal of translational medicine |
| 31807010 | FGFR2-BICC1: A Subtype Of FGFR2 Oncogenic Fusion Variant In Cholangiocarcinoma And The Response To Sorafenib. | Xihui Ying | 2019-01-01 | OncoTargets and therapy |
| 36071330 | Stochastic epigenetic mutations as possible explanation for phenotypical discordance among twins with congenital hypothyroidism. | D Gentilini | 2023-02-01 | Journal of endocrinological investigation |
| 32616843 | Induced pluripotent stem cells of patients with Tetralogy of Fallot reveal transcriptional alterations in cardiomyocyte differentiation. | Marcel Grunert | 2020-07-02 | Scientific reports |
| 30697050 | Serum BICC1 levels are significantly different in various mood disorders. | Suzhen Chen | 2019-01-01 | Neuropsychiatric disease and treatment |
| 31271100 | Protein Translation and Psychiatric Disorders. | Sophie Laguesse | 2020-02-01 | The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry |
| 33268819 | Fibroblast growth factor receptors in cancer: genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. | A Melanie Krook | 2021-03-01 | British journal of cancer |
| 31903140 | Mdig promotes oncogenic gene expression through antagonizing repressive histone methylation markers. | Qian Zhang | 2020-01-01 | Theranostics |
| 36660425 | An integrated analysis of prognostic mRNA signature in early- and progressive-stage gastric adenocarcinoma. | Xiaoling Hong | 2022-01-01 | Frontiers in molecular biosciences |
| 32252259 | Generation of An Endogenous FGFR2-BICC1 Gene Fusion/58 Megabase Inversion Using Single-Plasmid CRISPR/Cas9 Editing in Biliary Cells. | Andreas Reicher | 2020-04-02 | International journal of molecular sciences |
| 33977200 | Prioritization of Osteoporosis-Associated Genome-wide Association Study (GWAS) Single-Nucleotide Polymorphisms (SNPs) Using Epigenomics and Transcriptomics. | Xiao Zhang | 2021-05-01 | JBMR plus |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| BICC1-202 | ENST00000373886 | 5741 | 974aa | ENSP00000362993 |
| BICC1-203 | ENST00000476684 | 813 | No protein | - |
| BICC1-201 | ENST00000263103 | 1763 | 506aa | ENSP00000263103 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 28408 | Nutritional and Metabolic Diseases | 2.8720000E-005 | - |
| 28418 | Nutritional and Metabolic Diseases | 4.3450000E-005 | - |
| 28421 | Nutritional and Metabolic Diseases | 4.1370000E-005 | - |
| 28437 | Nutritional and Metabolic Diseases | 6.2200000E-005 | - |
| 64798 | Colorectal Neoplasms | 2E-7 | 26965516 |
| 65648 | Body Height | 7E-6 | 23251661 |
| 66507 | Circadian Rhythm | 9E-6 | 26835600 |
| 81940 | Myopia | 3E-14 | 27182965 |
| 82317 | Colorectal Neoplasms | 7E-8 | 24836286 |
| 93825 | Schizophrenia | 2E-6 | 26198764 |
| 97668 | Education | 3E-7 | 27020472 |
| 97669 | Refractive Errors | 3E-7 | 27020472 |
| 102961 | Education | 3E-9 | 27020472 |
| 102962 | Refractive Errors | 3E-9 | 27020472 |
| 106005 | Astigmatism | 3E-6 | 23322567 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
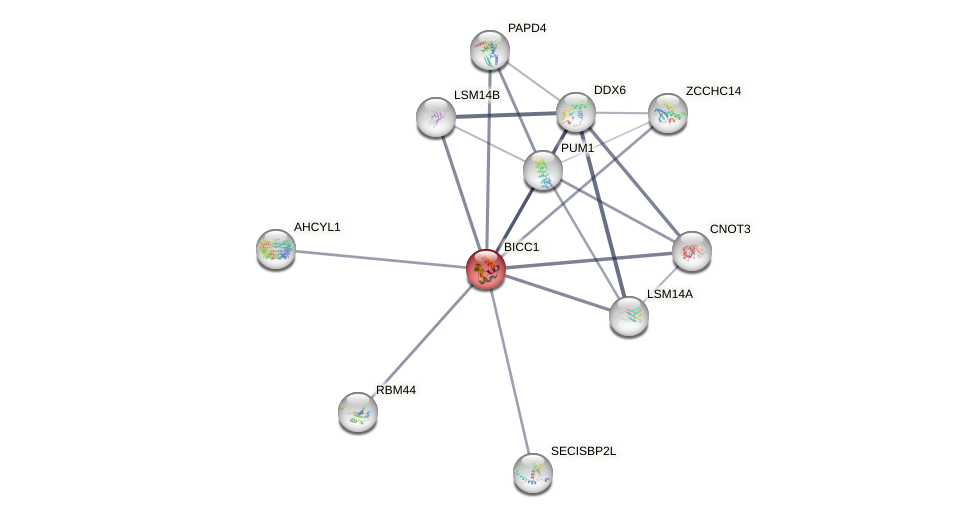
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000122870 | homo_sapiens | ENSP00000297837 | 34.4432 | 19.0586 | homo_sapiens | ENSP00000362993 | 30.8008 | 17.0431 |
| ENSG00000122870 | homo_sapiens | ENSP00000312042 | 28.0757 | 13.959 | homo_sapiens | ENSP00000362993 | 36.5503 | 18.1725 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 99.5893 | 99.076 | nomascus_leucogenys | ENSNLEP00000014920 | 99.5893 | 99.076 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.3573 | 98.2546 | pan_paniscus | ENSPPAP00000023935 | 99.8957 | 99.7915 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 99.4867 | 99.2813 | pongo_abelii | ENSPPYP00000038812 | 99.4867 | 99.2813 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 99.8973 | 99.7947 | pan_troglodytes | ENSPTRP00000074988 | 90.5959 | 90.5028 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 99.8973 | 99.7947 | gorilla_gorilla | ENSGGOP00000005374 | 99.8973 | 99.7947 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 92.7105 | 88.7064 | ornithorhynchus_anatinus | ENSOANP00000020891 | 91.8616 | 87.8942 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 78.7474 | 74.0246 | sarcophilus_harrisii | ENSSHAP00000013351 | 83.1887 | 78.1996 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 75.3593 | 71.9713 | notamacropus_eugenii | ENSMEUP00000005009 | 88.0096 | 84.0528 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 86.4476 | 84.2916 | choloepus_hoffmanni | ENSCHOP00000009594 | 86.8937 | 84.7265 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 89.7331 | 88.3984 | dasypus_novemcinctus | ENSDNOP00000007238 | 97.8723 | 96.4166 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 69.4045 | 66.8378 | erinaceus_europaeus | ENSEEUP00000001142 | 74.4493 | 71.696 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 87.9877 | 85.5236 | echinops_telfairi | ENSETEP00000008736 | 88.1687 | 85.6996 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 99.692 | 99.1786 | callithrix_jacchus | ENSCJAP00000019690 | 90.4097 | 89.9441 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 99.4867 | 99.384 | cercocebus_atys | ENSCATP00000004937 | 99.4867 | 99.384 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 99.5893 | 99.384 | macaca_fascicularis | ENSMFAP00000039353 | 85.3122 | 85.1363 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 99.5893 | 99.384 | macaca_mulatta | ENSMMUP00000020681 | 85.2373 | 85.0615 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 99.5893 | 99.384 | macaca_nemestrina | ENSMNEP00000002101 | 99.5893 | 99.384 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 94.0452 | 93.8398 | papio_anubis | ENSPANP00000020501 | 84.5018 | 84.3173 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.9733 | 98.768 | mandrillus_leucophaeus | ENSMLEP00000031633 | 99.484 | 99.2776 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 89.6304 | 88.6037 | tursiops_truncatus | ENSTTRP00000016406 | 96.0396 | 94.9395 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.46 | 97.3306 | vulpes_vulpes | ENSVVUP00000011336 | 98.46 | 97.3306 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 96.3039 | 93.1211 | mus_spretus | MGP_SPRETEiJ_P0024464 | 96.0082 | 92.8352 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.3573 | 97.8439 | equus_asinus | ENSEASP00005036419 | 89.6165 | 89.1487 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.1519 | 97.0226 | capra_hircus | ENSCHIP00000005699 | 98.3539 | 97.2222 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 87.7823 | 86.5503 | loxodonta_africana | ENSLAFP00000006509 | 98.0505 | 96.6743 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 97.9466 | 97.0226 | panthera_leo | ENSPLOP00000029643 | 97.9466 | 97.0226 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.3573 | 97.3306 | ailuropoda_melanoleuca | ENSAMEP00000033007 | 95.2286 | 94.2346 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 97.9466 | 96.7146 | bos_indicus_hybrid | ENSBIXP00005017989 | 88.8268 | 87.7095 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 93.5318 | 91.2731 | oryctolagus_cuniculus | ENSOCUP00000045700 | 92.7699 | 90.5295 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.3573 | 97.8439 | equus_caballus | ENSECAP00000068118 | 88.1325 | 87.6725 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 94.9692 | 93.5318 | canis_lupus_familiaris | ENSCAFP00845002790 | 88.0952 | 86.7619 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 97.9466 | 97.0226 | panthera_pardus | ENSPPRP00000001616 | 97.9466 | 97.0226 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.0493 | 96.8172 | marmota_marmota_marmota | ENSMMMP00000002636 | 98.0493 | 96.8172 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.2546 | 97.2279 | balaenoptera_musculus | ENSBMSP00010019948 | 98.2546 | 97.2279 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 91.5811 | 90.7598 | cavia_porcellus | ENSCPOP00000014053 | 97.6999 | 96.8237 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 97.8439 | 96.9199 | felis_catus | ENSFCAP00000005772 | 89.2322 | 88.3895 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 43.3265 | 40.6571 | monodelphis_domestica | ENSMODP00000020139 | 89.4068 | 83.8983 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 95.4825 | 94.2505 | bison_bison_bison | ENSBBBP00000019240 | 96.3731 | 95.1295 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.1519 | 96.9199 | monodon_monoceros | ENSMMNP00015026204 | 98.1519 | 96.9199 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.0493 | 97.0226 | sus_scrofa | ENSSSCP00000037472 | 89.4195 | 88.4831 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.8706 | 98.3573 | microcebus_murinus | ENSMICP00000036807 | 99.0741 | 98.5597 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 96.0986 | 94.4558 | octodon_degus | ENSODEP00000000650 | 96.0986 | 94.4558 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 95.4825 | 92.6078 | jaculus_jaculus | ENSJJAP00000011939 | 95.3846 | 92.5128 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.2546 | 97.0226 | ovis_aries_rambouillet | ENSOARP00020027970 | 88.9405 | 87.8253 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 95.5852 | 94.2505 | ursus_maritimus | ENSUMAP00000008677 | 95 | 93.6735 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.8809 | 82.4435 | ochotona_princeps | ENSOPRP00000008974 | 89.879 | 88.3388 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.1519 | 97.0226 | delphinapterus_leucas | ENSDLEP00000009470 | 98.1519 | 97.0226 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 97.9466 | 96.9199 | physeter_catodon | ENSPCTP00005027215 | 98.1481 | 97.1193 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 96.4066 | 93.0185 | mus_caroli | MGP_CAROLIEiJ_P0023317 | 96.1105 | 92.7329 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 96.2012 | 93.0185 | mus_musculus | ENSMUSP00000123201 | 95.9058 | 92.7329 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.3573 | 96.8172 | dipodomys_ordii | ENSDORP00000023467 | 98.3573 | 96.8172 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 76.078 | 72.7926 | mustela_putorius_furo | ENSMPUP00000010355 | 87.2792 | 83.51 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 96.6119 | 96.2012 | aotus_nancymaae | ENSANAP00000009631 | 99.3664 | 98.944 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 99.4867 | 98.9733 | saimiri_boliviensis_boliviensis | ENSSBOP00000027199 | 99.4867 | 98.9733 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 63.039 | 61.9097 | vicugna_pacos | ENSVPAP00000007646 | 63.299 | 62.1649 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 99.384 | 99.1786 | chlorocebus_sabaeus | ENSCSAP00000003566 | 99.384 | 99.1786 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 59.7536 | 58.9322 | tupaia_belangeri | ENSTBEP00000010599 | 64.0969 | 63.2159 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 96.2012 | 93.3265 | mesocricetus_auratus | ENSMAUP00000020884 | 96.3001 | 93.4224 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.2546 | 97.2279 | phocoena_sinus | ENSPSNP00000014495 | 98.2546 | 97.2279 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.46 | 97.3306 | camelus_dromedarius | ENSCDRP00005016734 | 98.46 | 97.3306 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 96.4066 | 95.9959 | carlito_syrichta | ENSTSYP00000004143 | 96.4066 | 95.9959 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 97.0226 | 96.0986 | panthera_tigris_altaica | ENSPTIP00000004501 | 98.029 | 97.0954 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 95.8932 | 93.0185 | rattus_norvegicus | ENSRNOP00000000754 | 95.3061 | 92.449 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.9733 | 97.9466 | prolemur_simus | ENSPSMP00000034565 | 99.177 | 98.1481 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 96.0986 | 93.6345 | microtus_ochrogaster | ENSMOCP00000023441 | 96.1973 | 93.7307 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 90.5544 | 89.2197 | sorex_araneus | ENSSARP00000010207 | 97.2437 | 95.8104 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 95.1745 | 92.8131 | peromyscus_maniculatus_bairdii | ENSPEMP00000014229 | 96.462 | 94.0687 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 95.3799 | 94.0452 | bos_mutus | ENSBMUP00000030259 | 98.3069 | 96.9312 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 89.425 | 86.037 | mus_spicilegus | ENSMSIP00000034045 | 91.5878 | 88.1178 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 96.2012 | 93.9425 | cricetulus_griseus_chok1gshd | ENSCGRP00001019839 | 96.3001 | 94.0391 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 93.2238 | 92.1971 | pteropus_vampyrus | ENSPVAP00000007555 | 93.2238 | 92.1971 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 88.2957 | 87.0637 | myotis_lucifugus | ENSMLUP00000004721 | 96.6292 | 95.2809 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 89.3224 | 85.8316 | vombatus_ursinus | ENSVURP00010001651 | 95.186 | 91.4661 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.2546 | 97.3306 | rhinolophus_ferrumequinum | ENSRFEP00010033358 | 98.2546 | 97.3306 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 90.0411 | 88.9117 | neovison_vison | ENSNVIP00000022900 | 97.9888 | 96.7598 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 97.8439 | 96.4066 | bos_taurus | ENSBTAP00000058535 | 97.7436 | 96.3077 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 81.9302 | 80.3901 | heterocephalus_glaber_female | ENSHGLP00000007763 | 96.7273 | 94.9091 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 90.0411 | 89.0144 | rhinopithecus_bieti | ENSRBIP00000013016 | 95.4298 | 94.3417 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.46 | 97.3306 | canis_lupus_dingo | ENSCAFP00020034689 | 98.46 | 97.3306 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.6653 | 97.1253 | sciurus_vulgaris | ENSSVLP00005010609 | 98.6653 | 97.1253 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 95.0719 | 91.3758 | phascolarctos_cinereus | ENSPCIP00000039333 | 95.1696 | 91.4697 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.0595 | 82.2382 | procavia_capensis | ENSPCAP00000006798 | 83.3162 | 82.4923 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 96.8172 | 94.4558 | nannospalax_galili | ENSNGAP00000025767 | 96.9168 | 94.5529 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 86.2423 | 84.3942 | bos_grunniens | ENSBGRP00000039640 | 97.1098 | 95.0289 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 89.9384 | 89.117 | chinchilla_lanigera | ENSCLAP00000017744 | 97.3333 | 96.4444 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.3573 | 97.4333 | cervus_hanglu_yarkandensis | ENSCHYP00000029701 | 89.1993 | 88.3613 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.0493 | 97.0226 | catagonus_wagneri | ENSCWAP00000029303 | 98.0493 | 97.0226 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 95.3799 | 94.2505 | ictidomys_tridecemlineatus | ENSSTOP00000000148 | 98.3069 | 97.1429 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.2546 | 97.1253 | otolemur_garnettii | ENSOGAP00000000272 | 98.4568 | 97.3251 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 99.4867 | 98.9733 | cebus_imitator | ENSCCAP00000020561 | 99.4867 | 98.9733 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.3573 | 97.1253 | urocitellus_parryii | ENSUPAP00010000757 | 89.3657 | 88.2463 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.1519 | 97.0226 | moschus_moschiferus | ENSMMSP00000027321 | 98.3539 | 97.2222 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 93.5318 | 92.5051 | rhinopithecus_roxellana | ENSRROP00000004379 | 98.1681 | 97.0905 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 96.3039 | 93.2238 | mus_pahari | MGP_PahariEiJ_P0086485 | 96.0082 | 92.9376 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 98.5626 | 97.9466 | propithecus_coquereli | ENSPCOP00000031651 | 98.7654 | 98.1481 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 12.731 | 8.93224 | ciona_intestinalis | ENSCINP00000031373 | 51.4523 | 36.0996 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 67.6591 | 53.7988 | petromyzon_marinus | ENSPMAP00000006065 | 70.7841 | 56.2836 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 48.768 | 36.2423 | eptatretus_burgeri | ENSEBUP00000025990 | 62.8307 | 46.6931 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 79.8768 | 73.306 | callorhinchus_milii | ENSCMIP00000047513 | 78.9047 | 72.4138 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 81.1088 | 75.9754 | latimeria_chalumnae | ENSLACP00000011472 | 90.9091 | 85.1553 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 86.037 | 78.5421 | lepisosteus_oculatus | ENSLOCP00000007857 | 84.9037 | 77.5076 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 72.6899 | 63.2444 | clupea_harengus | ENSCHAP00000062832 | 78.5794 | 68.3685 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 85.1129 | 75.77 | clupea_harengus | ENSCHAP00000025582 | 83.9069 | 74.6964 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 73.8193 | 62.423 | danio_rerio | ENSDARP00000129403 | 75.288 | 63.6649 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 48.2546 | 42.2998 | danio_rerio | ENSDARP00000126585 | 80.895 | 70.9122 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 85.2156 | 76.2834 | carassius_auratus | ENSCARP00000060998 | 85.567 | 76.5979 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 85.4209 | 76.1807 | carassius_auratus | ENSCARP00000051878 | 85.5967 | 76.3374 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 75.0513 | 63.347 | carassius_auratus | ENSCARP00000039519 | 74.9744 | 63.2821 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 74.846 | 63.5524 | carassius_auratus | ENSCARP00000078181 | 76.5756 | 65.021 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 85.4209 | 77.2074 | astyanax_mexicanus | ENSAMXP00000037005 | 85.5087 | 77.2867 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 63.347 | 53.1828 | astyanax_mexicanus | ENSAMXP00000011727 | 72.2482 | 60.6557 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 84.2916 | 76.078 | ictalurus_punctatus | ENSIPUP00000012412 | 82.018 | 74.026 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.8542 | 74.3326 | esox_lucius | ENSELUP00000028846 | 81.2689 | 72.9104 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 80.9035 | 70.2259 | esox_lucius | ENSELUP00000037859 | 71.6364 | 62.1818 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.8542 | 74.1273 | oncorhynchus_kisutch | ENSOKIP00005078815 | 81.5152 | 72.9293 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.9569 | 74.23 | oncorhynchus_kisutch | ENSOKIP00005100535 | 81.4516 | 72.8831 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 81.0062 | 71.9713 | oncorhynchus_mykiss | ENSOMYP00000121568 | 75.3582 | 66.9532 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.9569 | 74.23 | oncorhynchus_mykiss | ENSOMYP00000093977 | 81.6162 | 73.0303 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.8542 | 74.23 | salmo_salar | ENSSSAP00000081240 | 82.0956 | 73.5504 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 78.3368 | 69.5072 | salmo_salar | ENSSSAP00000003805 | 82.1313 | 72.8741 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.1622 | 74.23 | salmo_trutta | ENSSTUP00000107746 | 81.8182 | 73.0303 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.7515 | 74.1273 | salmo_trutta | ENSSTUP00000058922 | 81.9939 | 73.4486 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.2649 | 73.922 | gadus_morhua | ENSGMOP00000046862 | 82.335 | 73.0964 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 77.3101 | 67.4538 | fundulus_heteroclitus | ENSFHEP00000020472 | 82.9295 | 72.3568 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 78.2341 | 69.3018 | poecilia_reticulata | ENSPREP00000016029 | 83.4611 | 73.9321 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.8542 | 73.1006 | xiphophorus_maculatus | ENSXMAP00000034891 | 83.2817 | 73.4778 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 81.8275 | 73.1006 | oryzias_latipes | ENSORLP00000006699 | 82.7622 | 73.9356 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 37.0637 | 31.9302 | cyclopterus_lumpus | ENSCLMP00005040375 | 74.5868 | 64.2562 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.1622 | 74.4353 | oreochromis_niloticus | ENSONIP00000000788 | 83.4192 | 74.6653 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.8542 | 73.8193 | haplochromis_burtoni | ENSHBUP00000030776 | 83.1102 | 74.0474 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.8542 | 73.7166 | astatotilapia_calliptera | ENSACLP00000028682 | 83.1102 | 73.9444 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.7782 | 75.0513 | sparus_aurata | ENSSAUP00010068236 | 83.6066 | 74.8975 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 90.1437 | 83.3676 | xenopus_tropicalis | ENSXETP00000011134 | 90.2364 | 83.4532 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 92.9158 | 89.117 | chrysemys_picta_bellii | ENSCPBP00000007377 | 92.4413 | 88.6619 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 43.8398 | 41.1704 | sphenodon_punctatus | ENSSPUP00000004456 | 87.8601 | 82.5103 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 89.8357 | 85.6263 | crocodylus_porosus | ENSCPRP00005015196 | 84.5411 | 80.5797 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.7515 | 76.4887 | laticauda_laticaudata | ENSLLTP00000022742 | 86.8534 | 80.2802 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 88.7064 | 83.0595 | notechis_scutatus | ENSNSUP00000028257 | 77.9783 | 73.0144 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 89.117 | 83.2649 | pseudonaja_textilis | ENSPTXP00000025703 | 80.9701 | 75.653 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 85.5236 | 81.5195 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000015052 | 88.806 | 84.6482 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 93.2238 | 89.3224 | gallus_gallus | ENSGALP00010013845 | 90.8909 | 87.0871 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 84.9076 | 80.9035 | meleagris_gallopavo | ENSMGAP00000031199 | 90.6798 | 86.4035 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 85.9343 | 81.9302 | serinus_canaria | ENSSCAP00000004454 | 93.5196 | 89.162 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 92.1971 | 87.4743 | parus_major | ENSPMJP00000020756 | 91.7262 | 87.0276 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 91.4784 | 86.037 | anolis_carolinensis | ENSACAP00000001826 | 80.3427 | 75.5636 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.1622 | 74.3326 | neolamprologus_brichardi | ENSNBRP00000020018 | 83.7642 | 74.8707 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 90.1437 | 84.3942 | naja_naja | ENSNNAP00000027849 | 89.6833 | 83.9632 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 91.9918 | 87.269 | podarcis_muralis | ENSPMRP00000009681 | 76.7123 | 72.774 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 85.8316 | 81.7248 | geospiza_fortis | ENSGFOP00000016587 | 93.4078 | 88.9385 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 92.6078 | 88.3984 | taeniopygia_guttata | ENSTGUP00000031758 | 85.3359 | 81.457 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 85.1129 | 77.5154 | erpetoichthys_calabaricus | ENSECRP00000003212 | 85.7291 | 78.0765 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.7515 | 73.4086 | kryptolebias_marmoratus | ENSKMAP00000001081 | 83.0928 | 73.7113 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 91.7864 | 87.0637 | salvator_merianae | ENSSMRP00000021105 | 79.5374 | 75.4448 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.0595 | 73.8193 | oryzias_javanicus | ENSOJAP00000011641 | 83.1449 | 73.8952 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 42.5051 | 37.885 | amphilophus_citrinellus | ENSACIP00000008588 | 81.0176 | 72.2113 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.4702 | 74.846 | dicentrarchus_labrax | ENSDLAP00005075735 | 79.7059 | 71.4706 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.6756 | 74.9487 | amphiprion_percula | ENSAPEP00000008325 | 84.0206 | 75.2577 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 81.6222 | 72.7926 | oryzias_sinensis | ENSOSIP00000045172 | 82.1281 | 73.2438 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 84.9076 | 81.3142 | pelodiscus_sinensis | ENSPSIP00000010975 | 92.2991 | 88.3929 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 41.3758 | 38.0903 | gopherus_evgoodei | ENSGEVP00005022777 | 85.7447 | 78.9362 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 42.1971 | 38.9117 | chelonoidis_abingdonii | ENSCABP00000028874 | 86.8922 | 80.1268 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.5729 | 74.846 | seriola_dumerili | ENSSDUP00000023518 | 83.4016 | 74.6926 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.2649 | 74.6407 | cottoperca_gobio | ENSCGOP00000022244 | 83.1795 | 74.5641 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 88.3984 | 84.3942 | struthio_camelus_australis | ENSSCUP00000000210 | 92.9806 | 88.7689 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 93.2238 | 89.2197 | anser_brachyrhynchus | ENSABRP00000026574 | 92.8425 | 88.8548 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 79.5688 | 71.0472 | gasterosteus_aculeatus | ENSGACP00000013276 | 79.8146 | 71.2667 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.3676 | 74.3326 | cynoglossus_semilaevis | ENSCSEP00000007839 | 82.6884 | 73.7271 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.1622 | 73.306 | poecilia_formosa | ENSPFOP00000018482 | 83.5913 | 73.6842 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.9836 | 75.77 | myripristis_murdjan | ENSMMDP00005044177 | 84.0699 | 75.8479 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 93.1211 | 89.117 | strigops_habroptila | ENSSHBP00005010690 | 92.551 | 88.5714 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 81.6222 | 71.8686 | amphiprion_ocellaris | ENSAOCP00000030978 | 79.8995 | 70.3518 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 85.8316 | 82.3409 | terrapene_carolina_triunguis | ENSTMTP00000000972 | 93.3036 | 89.5089 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.9569 | 74.4353 | hucho_hucho | ENSHHUP00000061285 | 82.0305 | 73.6041 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.0329 | 73.306 | hucho_hucho | ENSHHUP00000072931 | 83.4029 | 74.5303 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 85.8316 | 77.0021 | pygocentrus_nattereri | ENSPNAP00000013124 | 85.9198 | 77.0812 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 65.1951 | 55.4415 | pygocentrus_nattereri | ENSPNAP00000019950 | 73.4954 | 62.5 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.3676 | 74.6407 | anabas_testudineus | ENSATEP00000062163 | 83.1967 | 74.4877 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 57.0842 | 41.6838 | ciona_savignyi | ENSCSAVP00000008102 | 63.1101 | 46.084 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.5462 | 73.8193 | seriola_lalandi_dorsalis | ENSSLDP00000020025 | 84.2767 | 75.3669 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.3676 | 74.846 | labrus_bergylta | ENSLBEP00000027968 | 83.1967 | 74.6926 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.8542 | 73.614 | pundamilia_nyererei | ENSPNYP00000003681 | 83.1102 | 73.8414 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.9836 | 74.846 | mastacembelus_armatus | ENSMAMP00000055511 | 83.8115 | 74.6926 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.6756 | 74.846 | stegastes_partitus | ENSSPAP00000020341 | 83.8477 | 75 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.8542 | 74.0246 | oncorhynchus_tshawytscha | ENSOTSP00005038398 | 81.5152 | 72.8283 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.9569 | 74.23 | oncorhynchus_tshawytscha | ENSOTSP00005055645 | 81.8642 | 73.2523 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.3676 | 74.846 | acanthochromis_polyacanthus | ENSAPOP00000023056 | 83.7977 | 75.2322 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 86.2423 | 82.8542 | aquila_chrysaetos_chrysaetos | ENSACCP00020002229 | 93.75 | 90.067 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.3409 | 72.8953 | nothobranchius_furzeri | ENSNFUP00015050774 | 83.368 | 73.8046 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.7782 | 74.1273 | cyprinodon_variegatus | ENSCVAP00000028651 | 84.0371 | 74.3563 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 80.5955 | 71.4579 | denticeps_clupeoides | ENSDCDP00000006595 | 78.6573 | 69.7395 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 63.1417 | 51.7454 | denticeps_clupeoides | ENSDCDP00000047184 | 75.0916 | 61.5385 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.9836 | 75.154 | denticeps_clupeoides | ENSDCDP00000012363 | 83.2146 | 74.4659 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.7782 | 74.9487 | larimichthys_crocea | ENSLCRP00005056763 | 83.6066 | 74.7951 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 81.6222 | 72.5873 | takifugu_rubripes | ENSTRUP00000021452 | 81.9588 | 72.8866 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 85.5236 | 81.5195 | ficedula_albicollis | ENSFALP00000002339 | 93.2811 | 88.9138 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.4435 | 73.922 | hippocampus_comes | ENSHCOP00000014095 | 82.4435 | 73.922 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 85.6263 | 76.694 | cyprinus_carpio_carpio | ENSCCRP00000107977 | 85.7143 | 76.7729 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 85.3183 | 76.5914 | cyprinus_carpio_carpio | ENSCCRP00000168743 | 82.8514 | 74.3769 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 74.23 | 62.731 | cyprinus_carpio_carpio | ENSCCRP00000037477 | 75.5486 | 63.8453 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 75.154 | 63.5524 | cyprinus_carpio_carpio | ENSCCRP00000124809 | 73.716 | 62.3364 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 93.2238 | 89.2197 | coturnix_japonica | ENSCJPP00005014782 | 85.4991 | 81.8267 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 83.3676 | 73.5113 | poecilia_latipinna | ENSPLAP00000007886 | 83.8843 | 73.9669 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 54.4148 | 45.3799 | sinocyclocheilus_grahami | ENSSGRP00000095000 | 72.7023 | 60.631 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 79.5688 | 70.5339 | sinocyclocheilus_grahami | ENSSGRP00000062184 | 85.5408 | 75.8278 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 80.3901 | 72.2793 | sinocyclocheilus_grahami | ENSSGRP00000004260 | 85.0163 | 76.4387 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 63.963 | 53.4908 | sinocyclocheilus_grahami | ENSSGRP00000078026 | 76.6298 | 64.0836 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.8542 | 73.7166 | maylandia_zebra | ENSMZEP00005036800 | 83.1102 | 73.9444 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 76.5914 | 66.3244 | tetraodon_nigroviridis | ENSTNIP00000020156 | 80.1289 | 69.3878 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 80.9035 | 70.7392 | tetraodon_nigroviridis | ENSTNIP00000022580 | 81.2371 | 71.0309 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 84.8049 | 77.4127 | paramormyrops_kingsleyae | ENSPKIP00000010671 | 84.7179 | 77.3333 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.7515 | 75.5647 | scleropages_formosus | ENSSFOP00015017903 | 81.5789 | 74.4939 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 74.9487 | 65.9138 | scleropages_formosus | ENSSFOP00015013087 | 77.9915 | 68.5897 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 89.3224 | 82.8542 | leptobrachium_leishanense | ENSLLEP00000014008 | 88.0567 | 81.6802 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 79.0554 | 69.9179 | scophthalmus_maximus | ENSSMAP00000027870 | 78.7321 | 69.6319 |
| ENSG00000122870 | homo_sapiens | ENSP00000362993 | 82.9569 | 74.0246 | oryzias_melastigma | ENSOMEP00000027269 | 83.3849 | 74.4066 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0001822 | involved_in kidney development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0005737 | is_active_in cytoplasm | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0007368 | involved_in determination of left/right symmetry | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007507 | involved_in heart development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0090090 | involved_in negative regulation of canonical Wnt signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |