RBP Type
Non-canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
DUB
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000121022 Gene ID
10987 Accession
2240
Symbol
COPS5
Alias
CSN5;JAB1;SGN5;MOV-34
Full Name
COP9 signalosome subunit 5
Status
Confidence
Length
21865 bases
Strand
Minus strand
Position
8 : 67043079 - 67064943
RNA binding domain
N.A.
Summary
The protein encoded by this gene is one of the eight subunits of COP9 signalosome, a highly conserved protein complex that functions as an important regulator in multiple signaling pathways. The structure and function of COP9 signalosome is similar to that of the 19S regulatory particle of 26S proteasome. COP9 signalosome has been shown to interact with SCF-type E3 ubiquitin ligases and act as a positive regulator of E3 ubiquitin ligases. This protein is reported to be involved in the degradation of cyclin-dependent kinase inhibitor CDKN1B/p27Kip1. It is also known to be an coactivator that increases the specificity of JUN/AP1 transcription factors. [provided by RefSeq, Jul 2008]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
27903243 Analysis of RNA expression of normal and cancer tissues reveals high correlation of COP9 gene expression with respiratory chain complex components.
A Christina Wicker
2016-12-01
BMC genomics
Name
Transcript ID
bp
Protein
Translation ID
COPS5-212
ENST00000523086
717
No protein
-
COPS5-201
ENST00000357849
1296
334aa
ENSP00000350512
COPS5-211
ENST00000521509
3826
No protein
-
COPS5-203
ENST00000517736
1621
197aa
ENSP00000429774
COPS5-210
ENST00000521386
5691
No protein
-
COPS5-205
ENST00000518374
1451
68aa
ENSP00000427869
COPS5-213
ENST00000523890
1086
57aa
ENSP00000427997
COPS5-206
ENST00000518747
676
151aa
ENSP00000428586
COPS5-204
ENST00000517793
657
No protein
-
COPS5-202
ENST00000517406
474
78aa
ENSP00000428124
COPS5-209
ENST00000519963
764
No protein
-
COPS5-208
ENST00000519057
566
No protein
-
COPS5-207
ENST00000518768
565
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
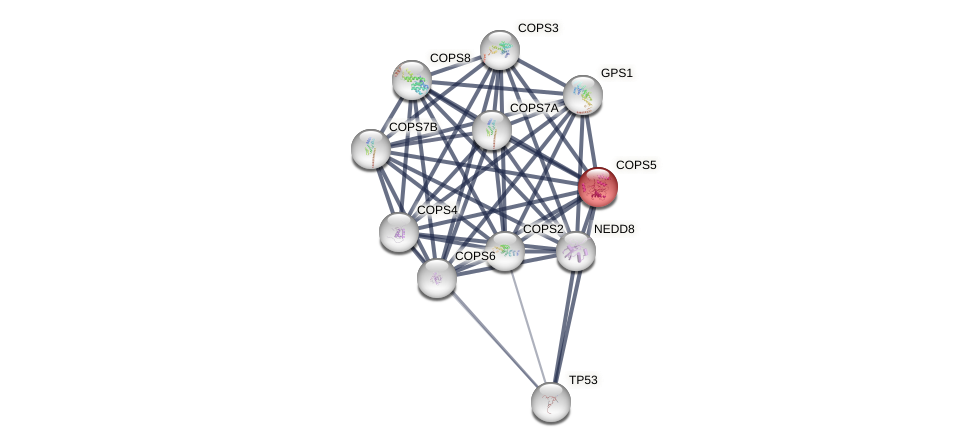
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000338 involved_in protein deneddylation 3 IBA,IDA,IMP Homo_sapiens(9606) Process GO:0003713 enables transcription coactivator activity 1 TAS Homo_sapiens(9606) Function GO:0003743 enables translation initiation factor activity 1 TAS Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 located_in nucleus 2 IDA,ISS Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 2 IDA,TAS Homo_sapiens(9606) Component GO:0005737 is_active_in cytoplasm 3 IBA,IDA,ISS Homo_sapiens(9606) Component GO:0005829 located_in cytosol 2 IDA,TAS Homo_sapiens(9606) Component GO:0005852 part_of eukaryotic translation initiation factor 3 complex 1 TAS Homo_sapiens(9606) Component GO:0006412 involved_in translation 1 TAS Homo_sapiens(9606) Process GO:0006413 involved_in translational initiation 1 IEA Homo_sapiens(9606) Process GO:0008021 located_in synaptic vesicle 1 IDA Homo_sapiens(9606) Component GO:0008180 part_of COP9 signalosome 4 IBA,IDA,IPI,ISS Homo_sapiens(9606) Component GO:0008237 enables metallopeptidase activity 2 IBA,IMP Homo_sapiens(9606) Function GO:0016579 involved_in protein deubiquitination 1 IDA Homo_sapiens(9606) Process GO:0019784 enables deNEDDylase activity 2 IBA,TAS Homo_sapiens(9606) Function GO:0019899 enables enzyme binding 1 IEA Homo_sapiens(9606) Function GO:0035718 enables macrophage migration inhibitory factor binding 1 IEA Homo_sapiens(9606) Function GO:0043066 involved_in negative regulation of apoptotic process 1 IMP Homo_sapiens(9606) Process GO:0043687 involved_in post-translational protein modification 1 TAS Homo_sapiens(9606) Process GO:0045116 involved_in protein neddylation 1 NAS Homo_sapiens(9606) Process GO:0045944 involved_in positive regulation of transcription by RNA polymerase II 1 IDA Homo_sapiens(9606) Process GO:0046328 involved_in regulation of JNK cascade 1 IDA Homo_sapiens(9606) Process GO:0046872 enables metal ion binding 1 IEA Homo_sapiens(9606) Function GO:0048471 located_in perinuclear region of cytoplasm 1 IDA Homo_sapiens(9606) Component GO:0051091 involved_in positive regulation of DNA-binding transcription factor activity 1 IDA Homo_sapiens(9606) Process GO:0051726 involved_in regulation of cell cycle 1 IEA Homo_sapiens(9606) Process GO:0140492 enables metal-dependent deubiquitinase activity 1 IDA Homo_sapiens(9606) Function GO:1903894 involved_in regulation of IRE1-mediated unfolded protein response 1 IMP Homo_sapiens(9606) Process GO:1990182 involved_in exosomal secretion 1 IDA Homo_sapiens(9606) Process GO:2000434 involved_in regulation of protein neddylation 1 NAS Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.