RBP Type
Canonical_RBPs
Diseases
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
N.A.
Localizations
N.A.
BulkPerturb-seq
N.A.
Ensembl ID ENSG00000108387 Gene ID
5414 Accession
9165
Symbol
SEPTIN4
Alias
H5;ARTS;MART;SEP4;CE5B3;SEPT4;PNUTL2;hucep-7;BRADEION;C17orf47;hCDCREL-2
Full Name
septin 4
Status
Confidence
Length
24119 bases
Strand
Minus strand
Position
17 : 58520250 - 58544368
RNA binding domain
MMR_HSR1
Summary
This gene is a member of the septin family of nucleotide binding proteins, originally described in yeast as cell division cycle regulatory proteins. Septins are highly conserved in yeast, Drosophila, and mouse, and appear to regulate cytoskeletal organization. Disruption of septin function disturbs cytokinesis and results in large multinucleate or polyploid cells. This gene is highly expressed in brain and heart. Alternatively spliced transcript variants encoding different isoforms have been described for this gene. One of the isoforms (known as ARTS) is distinct; it is localized to the mitochondria, and has a role in apoptosis and cancer. [provided by RefSeq, Nov 2010]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000463671
MMR_HSR1
PF01926 9.8e-07
1
ENSP00000464373
MMR_HSR1
PF01926 2.3e-06
1
ENSP00000462727
MMR_HSR1
PF01926 2.4e-06
1
ENSP00000463768
MMR_HSR1
PF01926 3.6e-06
1
ENSP00000321071
MMR_HSR1
PF01926 3.9e-06
1
ENSP00000376801
MMR_HSR1
PF01926 3.9e-06
1
ENSP00000414779
MMR_HSR1
PF01926 4e-06
1
ENSP00000321674
MMR_HSR1
PF01926 4.3e-06
1
ENSP00000402000
MMR_HSR1
PF01926 4.5e-06
1
ENSP00000500355
MMR_HSR1
PF01926 4.5e-06
1
ENSP00000464382
MMR_HSR1
PF01926 5.2e-06
1
ENSP00000402348
MMR_HSR1
PF01926 1.5e-05
1
ENSP00000500383
MMR_HSR1
PF01926 1.6e-05
1
ENSP00000463461
MMR_HSR1
PF01926 5.7e-05
1
ENSP00000500629
MMR_HSR1
PF01926 6.3e-05
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
SEPTIN4-204
ENST00000393086
1556
459aa
ENSP00000376801
SEPTIN4-224
ENST00000583114
1709
331aa
ENSP00000463768
SEPTIN4-232
ENST00000672673
3303
996aa
ENSP00000500383
SEPTIN4-206
ENST00000426861
1906
274aa
ENSP00000402348
SEPTIN4-215
ENST00000580796
2790
No protein
-
SEPTIN4-222
ENST00000582270
2195
No protein
-
SEPTIN4-212
ENST00000579371
1362
332aa
ENSP00000464373
SEPTIN4-225
ENST00000583273
2151
No protein
-
SEPTIN4-202
ENST00000317268
1784
478aa
ENSP00000321674
SEPTIN4-208
ENST00000577440
1792
No protein
-
SEPTIN4-207
ENST00000457347
1795
493aa
ENSP00000402000
SEPTIN4-233
ENST00000672699
1795
493aa
ENSP00000500355
SEPTIN4-217
ENST00000580844
1396
379aa
ENSP00000462727
SEPTIN4-201
ENST00000317256
1530
459aa
ENSP00000321071
SEPTIN4-205
ENST00000412945
1597
470aa
ENSP00000414779
SEPTIN4-227
ENST00000584488
1731
No protein
-
SEPTIN4-216
ENST00000580809
952
175aa
ENSP00000464382
SEPTIN4-209
ENST00000577729
724
207aa
ENSP00000463671
SEPTIN4-219
ENST00000581615
632
55aa
ENSP00000462278
SEPTIN4-210
ENST00000578131
508
36aa
ENSP00000462362
SEPTIN4-230
ENST00000585170
3544
No protein
-
SEPTIN4-226
ENST00000583291
789
225aa
ENSP00000463461
SEPTIN4-231
ENST00000671766
705
211aa
ENSP00000500629
SEPTIN4-229
ENST00000584789
963
No protein
-
SEPTIN4-220
ENST00000581921
235
No protein
-
SEPTIN4-228
ENST00000584528
559
No protein
-
SEPTIN4-214
ENST00000580791
566
No protein
-
SEPTIN4-221
ENST00000582248
604
No protein
-
SEPTIN4-213
ENST00000580740
549
No protein
-
SEPTIN4-211
ENST00000578747
564
No protein
-
SEPTIN4-218
ENST00000581607
676
72aa
ENSP00000464360
SEPTIN4-223
ENST00000582976
1707
No protein
-
SEPTIN4-203
ENST00000321691
2123
570aa
ENSP00000354874
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
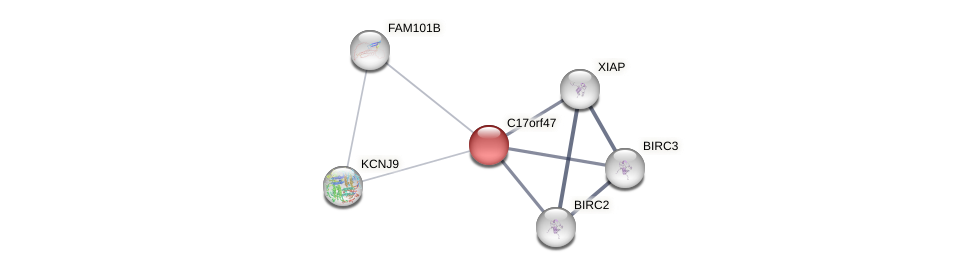
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000108387
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000108387 homo_sapiens ENSP00000500383 96.3855 95.3815 nomascus_leucogenys ENSNLEP00000010107 95.0495 94.0594 ENSG00000108387 homo_sapiens ENSP00000500383 98.8956 98.494 pan_paniscus ENSPPAP00000010095 97.332 96.9368 ENSG00000108387 homo_sapiens ENSP00000500383 98.3936 97.7912 pongo_abelii ENSPPYP00000009246 98.3936 97.7912 ENSG00000108387 homo_sapiens ENSP00000500383 98.7952 98.3936 pan_troglodytes ENSPTRP00000016057 97.2332 96.8379 ENSG00000108387 homo_sapiens ENSP00000500383 98.6948 98.2932 gorilla_gorilla ENSGGOP00000008193 97.1344 96.7391 ENSG00000108387 homo_sapiens ENSP00000500383 20.0803 13.8554 notamacropus_eugenii ENSMEUP00000004470 50.1253 34.5865 ENSG00000108387 homo_sapiens ENSP00000500383 37.0482 28.4137 erinaceus_europaeus ENSEEUP00000008759 67.2131 51.5483 ENSG00000108387 homo_sapiens ENSP00000500383 33.4337 26.004 echinops_telfairi ENSETEP00000013922 61.8959 48.1413 ENSG00000108387 homo_sapiens ENSP00000500383 46.4859 43.2731 callithrix_jacchus ENSCJAP00000081055 85.7407 79.8148 ENSG00000108387 homo_sapiens ENSP00000500383 92.2691 88.3534 callithrix_jacchus ENSCJAP00000087998 92.2691 88.3534 ENSG00000108387 homo_sapiens ENSP00000500383 94.3775 92.9719 cercocebus_atys ENSCATP00000018089 95.4315 94.0102 ENSG00000108387 homo_sapiens ENSP00000500383 96.7871 95.1807 macaca_fascicularis ENSMFAP00000038356 96.8844 95.2764 ENSG00000108387 homo_sapiens ENSP00000500383 52.6104 50.6024 macaca_mulatta ENSMMUP00000014448 92.0914 88.5764 ENSG00000108387 homo_sapiens ENSP00000500383 94.4779 92.9719 macaca_nemestrina ENSMNEP00000026249 95.533 94.0102 ENSG00000108387 homo_sapiens ENSP00000500383 96.7871 95.3815 papio_anubis ENSPANP00000039269 96.8844 95.4774 ENSG00000108387 homo_sapiens ENSP00000500383 94.4779 93.0723 mandrillus_leucophaeus ENSMLEP00000006780 95.533 94.1117 ENSG00000108387 homo_sapiens ENSP00000500383 37.9518 31.7269 tursiops_truncatus ENSTTRP00000016287 69.3578 57.9817 ENSG00000108387 homo_sapiens ENSP00000500383 39.257 33.0321 vulpes_vulpes ENSVVUP00000017218 72.4074 60.9259 ENSG00000108387 homo_sapiens ENSP00000500383 35.241 28.012 mus_spretus MGP_SPRETEiJ_P0030454 65.9774 52.4436 ENSG00000108387 homo_sapiens ENSP00000500383 85.743 79.0161 equus_asinus ENSEASP00005003628 84.9751 78.3085 ENSG00000108387 homo_sapiens ENSP00000500383 38.253 32.1285 loxodonta_africana ENSLAFP00000019502 73.8372 62.0155 ENSG00000108387 homo_sapiens ENSP00000500383 84.9398 78.2129 panthera_leo ENSPLOP00000016337 85.5409 78.7664 ENSG00000108387 homo_sapiens ENSP00000500383 84.2369 77.4096 ailuropoda_melanoleuca ENSAMEP00000012183 85.1777 78.2741 ENSG00000108387 homo_sapiens ENSP00000500383 33.0321 28.012 canis_lupus_familiaris ENSCAFP00845030057 72.1491 61.1842 ENSG00000108387 homo_sapiens ENSP00000500383 41.2651 34.8394 panthera_pardus ENSPPRP00000004660 73.7882 62.298 ENSG00000108387 homo_sapiens ENSP00000500383 82.4297 75.7028 balaenoptera_musculus ENSBMSP00010024938 84.552 77.6519 ENSG00000108387 homo_sapiens ENSP00000500383 39.3574 33.5341 felis_catus ENSFCAP00000008259 73.271 62.4299 ENSG00000108387 homo_sapiens ENSP00000500383 25.8032 18.0723 monodelphis_domestica ENSMODP00000048049 47.3297 33.1492 ENSG00000108387 homo_sapiens ENSP00000500383 40.5622 35.3414 microcebus_murinus ENSMICP00000014658 72.0143 62.7451 ENSG00000108387 homo_sapiens ENSP00000500383 33.6345 27.8112 octodon_degus ENSODEP00000017855 67.4044 55.7344 ENSG00000108387 homo_sapiens ENSP00000500383 35.241 28.6145 jaculus_jaculus ENSJJAP00000021476 70.9091 57.5758 ENSG00000108387 homo_sapiens ENSP00000500383 28.7149 23.8956 ochotona_princeps ENSOPRP00000014400 50.7993 42.2735 ENSG00000108387 homo_sapiens ENSP00000500383 35.4418 28.3133 mus_caroli MGP_CAROLIEiJ_P0029042 66.1049 52.809 ENSG00000108387 homo_sapiens ENSP00000500383 32.9317 26.1044 dipodomys_ordii ENSDORP00000027068 66.3968 52.6316 ENSG00000108387 homo_sapiens ENSP00000500383 83.9357 76.2048 mustela_putorius_furo ENSMPUP00000015175 83.2669 75.5976 ENSG00000108387 homo_sapiens ENSP00000500383 48.7952 46.5863 aotus_nancymaae ENSANAP00000030565 86.3233 82.4156 ENSG00000108387 homo_sapiens ENSP00000500383 49.498 46.6867 saimiri_boliviensis_boliviensis ENSSBOP00000007663 85 80.1724 ENSG00000108387 homo_sapiens ENSP00000500383 38.9558 32.7309 vicugna_pacos ENSVPAP00000001667 71.5867 60.1476 ENSG00000108387 homo_sapiens ENSP00000500383 52.1084 50.1004 chlorocebus_sabaeus ENSCSAP00000002086 91.0526 87.5439 ENSG00000108387 homo_sapiens ENSP00000500383 35.6426 29.7189 tupaia_belangeri ENSTBEP00000008912 63.1673 52.669 ENSG00000108387 homo_sapiens ENSP00000500383 35.1406 28.7149 mesocricetus_auratus ENSMAUP00000016463 69.8603 57.0858 ENSG00000108387 homo_sapiens ENSP00000500383 81.6265 74.7992 phocoena_sinus ENSPSNP00000024693 84.0745 77.0424 ENSG00000108387 homo_sapiens ENSP00000500383 41.4659 35.8434 carlito_syrichta ENSTSYP00000017350 73.8819 63.864 ENSG00000108387 homo_sapiens ENSP00000500383 41.2651 34.739 panthera_tigris_altaica ENSPTIP00000024383 73.7882 62.1185 ENSG00000108387 homo_sapiens ENSP00000500383 85.743 80.6225 prolemur_simus ENSPSMP00000004636 85.4855 80.3804 ENSG00000108387 homo_sapiens ENSP00000500383 29.8193 21.6867 sorex_araneus ENSSARP00000011566 59.4 43.2 ENSG00000108387 homo_sapiens ENSP00000500383 34.3373 27.4096 peromyscus_maniculatus_bairdii ENSPEMP00000028301 68.1275 54.3825 ENSG00000108387 homo_sapiens ENSP00000500383 35.6426 28.4137 mus_spicilegus ENSMSIP00000031786 66.855 53.2957 ENSG00000108387 homo_sapiens ENSP00000500383 35.3414 28.4137 cricetulus_griseus_chok1gshd ENSCGRP00001021011 70.2595 56.487 ENSG00000108387 homo_sapiens ENSP00000500383 40.3614 33.4337 pteropus_vampyrus ENSPVAP00000002201 71.0247 58.8339 ENSG00000108387 homo_sapiens ENSP00000500383 40.8635 33.7349 rhinolophus_ferrumequinum ENSRFEP00010019937 71.1538 58.7413 ENSG00000108387 homo_sapiens ENSP00000500383 51.506 48.8956 rhinopithecus_bieti ENSRBIP00000019956 90.4762 85.8907 ENSG00000108387 homo_sapiens ENSP00000500383 38.755 32.8313 canis_lupus_dingo ENSCAFP00020033423 71.4815 60.5556 ENSG00000108387 homo_sapiens ENSP00000500383 85.9438 79.5181 sciurus_vulgaris ENSSVLP00005016003 86.6397 80.1619 ENSG00000108387 homo_sapiens ENSP00000500383 33.9357 29.3173 ictidomys_tridecemlineatus ENSSTOP00000020134 71.308 61.6034 ENSG00000108387 homo_sapiens ENSP00000500383 40.261 33.6345 otolemur_garnettii ENSOGAP00000022406 71.0993 59.3972 ENSG00000108387 homo_sapiens ENSP00000500383 49.6988 46.6867 cebus_imitator ENSCCAP00000029372 85.1979 80.0344 ENSG00000108387 homo_sapiens ENSP00000500383 51.506 48.8956 rhinopithecus_roxellana ENSRROP00000034856 90.4762 85.8907 ENSG00000108387 homo_sapiens ENSP00000500383 35.0402 27.7108 mus_pahari MGP_PahariEiJ_P0035756 64.3911 50.9225 ENSG00000108387 homo_sapiens ENSP00000500383 43.9759 39.1566 propithecus_coquereli ENSPCOP00000013128 78.2143 69.6429
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000287 enables magnesium ion binding 1 IMP Homo_sapiens(9606) Function GO:0001764 involved_in neuron migration 1 ISS Homo_sapiens(9606) Process GO:0003924 enables GTPase activity 2 IBA,IMP Homo_sapiens(9606) Function GO:0005198 enables structural molecule activity 1 TAS Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005525 enables GTP binding 1 IMP Homo_sapiens(9606) Function GO:0005634 located_in nucleus 1 NAS Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 1 IDA Homo_sapiens(9606) Component GO:0005739 located_in mitochondrion 2 IDA,NAS Homo_sapiens(9606) Component GO:0005741 located_in mitochondrial outer membrane 1 TAS Homo_sapiens(9606) Component GO:0005829 located_in cytosol 1 TAS Homo_sapiens(9606) Component GO:0005940 is_active_in septin ring 1 IBA Homo_sapiens(9606) Component GO:0006915 involved_in apoptotic process 1 NAS Homo_sapiens(9606) Process GO:0008021 is_active_in synaptic vesicle 1 IBA Homo_sapiens(9606) Component GO:0015630 is_active_in microtubule cytoskeleton 1 IBA Homo_sapiens(9606) Component GO:0017157 involved_in regulation of exocytosis 1 IBA Homo_sapiens(9606) Process GO:0030317 involved_in flagellated sperm motility 1 ISS Homo_sapiens(9606) Process GO:0030424 located_in axon 1 ISS Homo_sapiens(9606) Component GO:0030425 located_in dendrite 1 ISS Homo_sapiens(9606) Component GO:0031105 part_of septin complex 3 IBA,IDA,IMP Homo_sapiens(9606) Component GO:0031398 involved_in positive regulation of protein ubiquitination 1 IDA Homo_sapiens(9606) Process GO:0032153 is_active_in cell division site 1 IBA Homo_sapiens(9606) Component GO:0042802 enables identical protein binding 1 IPI Homo_sapiens(9606) Function GO:0042981 involved_in regulation of apoptotic process 1 NAS Homo_sapiens(9606) Process GO:0043065 involved_in positive regulation of apoptotic process 1 IDA Homo_sapiens(9606) Process GO:0043204 located_in perikaryon 1 ISS Homo_sapiens(9606) Component GO:0048515 involved_in spermatid differentiation 1 ISS Homo_sapiens(9606) Process GO:0060090 enables molecular adaptor activity 1 IBA Homo_sapiens(9606) Function GO:0061484 involved_in hematopoietic stem cell homeostasis 1 ISS Homo_sapiens(9606) Process GO:0061640 involved_in cytoskeleton-dependent cytokinesis 1 IBA Homo_sapiens(9606) Process GO:0097227 is_active_in sperm annulus 2 IBA,IDA Homo_sapiens(9606) Component GO:2001244 involved_in positive regulation of intrinsic apoptotic signaling pathway 1 IMP Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.