| RBP Type |
Canonical_RBPs |
| Diseases |
FlavivirusesHIVSARS-COV-2SINVDengueZika |
| Drug |
N.A. |
| Main interacting RNAs |
mRNA |
| Moonlighting functions |
N.A. |
| Localizations |
Stress granucle |
|
BulkPerturb-seq
|
DataSet_01_285 DataSet_01_372 |
| Ensembl ID | ENSG00000108256 | Gene ID |
57532 |
Accession |
17634 |
| Symbol |
NUFIP2 |
Alias |
|
Full Name |
|
| Status |
Confidence |
Length |
38310 bases |
Strand |
Minus strand |
| Position |
17 : 29255839 - 29294148 |
RNA binding domain |
NUFIP2 |
| Summary |
|
RNA binding domains (RBDs)

| Protein |
Domain |
Pfam ID |
E-value |
Domain number |
| ENSP00000225388 |
NUFIP2 |
PF15293 |
0 |
1 |
RNA binding proteomes (RBPomes)
| Pubmed ID |
Full Name |
Cell |
Author |
Time |
Doi |
Literatures on RNA binding capacity
| Pubmed ID |
Title |
Author |
Time |
Journal |
| 29352114 |
Binding of NUFIP2 to Roquin promotes recognition and regulation of ICOS mRNA. |
Nina Rehage |
2018-01-19 |
Nature communications |
| 35467332 |
Writing and Erasing O-GlcNAc on Casein Kinase 2 Alpha Alters the Phosphoproteome. |
A Paul Schwein |
2022-05-20 |
ACS chemical biology |
| 34604665 |
Interactome Networks of FOSL1 and FOSL2 in Human Th17 Cells. |
Ankitha Shetty |
2021-09-28 |
ACS omega |
| 37491482 |
Plasma microRNA ratios associated with breast cancer detection in a nested case-control study from a mammography screening cohort. |
Giovanna Chiorino |
2023-07-25 |
Scientific reports |
| 26184334 |
Comprehensive Protein Interactome Analysis of a Key RNA Helicase: Detection of Novel Stress Granule Proteins. |
Rebecca Bish |
2015-07-15 |
Biomolecules |
| Name |
Transcript ID |
bp |
Protein |
Translation ID |
| NUFIP2-201 |
ENST00000225388 |
10877 |
695aa |
ENSP00000225388 |
| NUFIP2-202 |
ENST00000579665 |
559 |
120aa |
ENSP00000463450 |
| Pathway ID |
Pathway Name |
Source |
| | |
| ensgID |
Trait |
pValue |
Pubmed ID |
| 2170 | Body Mass Index | 7.65346517690446E-5 | 17903300 |
| 4712 | Cholesterol, HDL | 4.75951132718611E-7 | 17903299 |
| ensgID | SNP | Chromosome | Position |
Trait | PubmedID | Or or BEAT |
EFO ID |
|---|
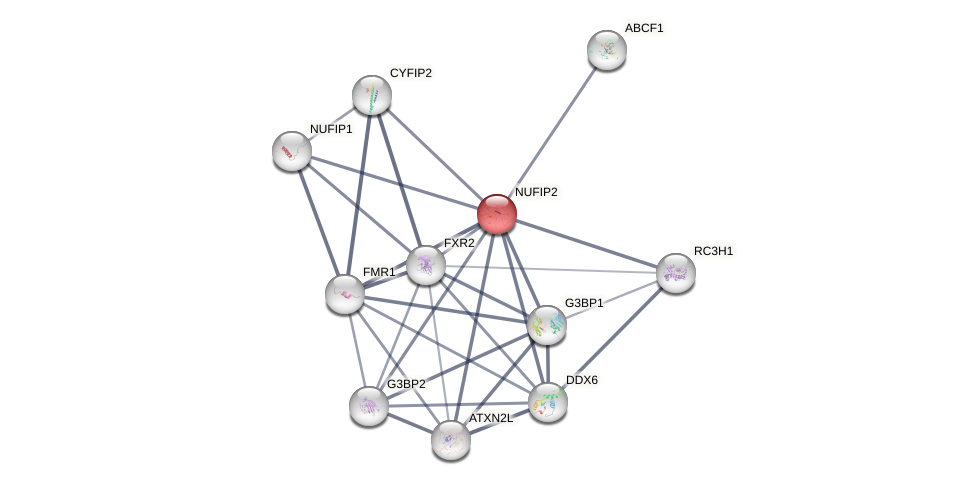
Protein-Protein Interaction (PPI)
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSG00000108256 | | | | | | | | |
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.4245 | 98.9928 | nomascus_leucogenys | ENSNLEP00000003996 | 99.5677 | 99.1354 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.8561 | 99.8561 | pan_paniscus | ENSPPAP00000013929 | 99.8561 | 99.8561 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.7122 | 99.5683 | pongo_abelii | ENSPPYP00000009131 | 99.7122 | 99.5683 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.7122 | 99.7122 | pan_troglodytes | ENSPTRP00000015274 | 99.7122 | 99.7122 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.8561 | 99.8561 | gorilla_gorilla | ENSGGOP00000008396 | 99.8561 | 99.8561 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 88.3453 | 80.1439 | ornithorhynchus_anatinus | ENSOANP00000020002 | 88.4726 | 80.2594 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 90.5036 | 84.1727 | sarcophilus_harrisii | ENSSHAP00000019358 | 65.7262 | 61.1285 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 56.259 | 51.0791 | notamacropus_eugenii | ENSMEUP00000014973 | 56.259 | 51.0791 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 96.8345 | 93.3813 | choloepus_hoffmanni | ENSCHOP00000003002 | 97.114 | 93.6508 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 58.4173 | 56.5468 | choloepus_hoffmanni | ENSCHOP00000002425 | 58.5859 | 56.71 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.1223 | 94.3885 | dasypus_novemcinctus | ENSDNOP00000011250 | 94.0111 | 91.3649 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 81.4389 | 76.6907 | erinaceus_europaeus | ENSEEUP00000007702 | 94.4908 | 88.9816 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 92.3741 | 88.4892 | echinops_telfairi | ENSETEP00000002714 | 95.5357 | 91.5179 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.1367 | 97.9856 | callithrix_jacchus | ENSCJAP00000082711 | 99.1367 | 97.9856 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.5683 | 98.9928 | cercocebus_atys | ENSCATP00000014519 | 99.5683 | 98.9928 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.7122 | 99.1367 | macaca_fascicularis | ENSMFAP00000042995 | 99.7122 | 99.1367 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.7122 | 98.9928 | macaca_mulatta | ENSMMUP00000016889 | 99.7122 | 98.9928 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.4101 | 95.5396 | macaca_nemestrina | ENSMNEP00000030919 | 96.3016 | 94.4523 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.4245 | 98.8489 | papio_anubis | ENSPANP00000019909 | 99.4245 | 98.8489 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.5683 | 98.9928 | mandrillus_leucophaeus | ENSMLEP00000024597 | 99.5683 | 98.9928 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 81.5827 | 78.9928 | tursiops_truncatus | ENSTTRP00000015476 | 81.7003 | 79.1066 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.6978 | 94.964 | vulpes_vulpes | ENSVVUP00000028707 | 97.2779 | 94.5559 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.1223 | 93.9568 | mus_spretus | MGP_SPRETEiJ_P0029833 | 92.4658 | 89.4521 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.4173 | 96.1151 | equus_asinus | ENSEASP00005014055 | 98.4173 | 96.1151 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.2734 | 94.964 | capra_hircus | ENSCHIP00000014771 | 98.2734 | 94.964 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 94.964 | 92.3741 | loxodonta_africana | ENSLAFP00000025514 | 97.0588 | 94.4118 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.8417 | 95.2518 | panthera_leo | ENSPLOP00000009455 | 96.0452 | 93.5028 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.4173 | 95.3957 | ailuropoda_melanoleuca | ENSAMEP00000004564 | 97.4359 | 94.4444 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.5611 | 95.2518 | bos_indicus_hybrid | ENSBIXP00005034535 | 98.2783 | 94.9785 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.4173 | 95.9712 | oryctolagus_cuniculus | ENSOCUP00000014169 | 98.2759 | 95.8333 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.4173 | 96.259 | equus_caballus | ENSECAP00000003540 | 98.2759 | 96.1207 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 96.4029 | 93.0935 | canis_lupus_familiaris | ENSCAFP00845015153 | 93.185 | 89.9861 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 88.777 | 86.187 | panthera_pardus | ENSPPRP00000008949 | 98.0922 | 95.2305 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 95.5396 | 92.2302 | marmota_marmota_marmota | ENSMMMP00000003694 | 95.2654 | 91.9656 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.2662 | 94.8201 | balaenoptera_musculus | ENSBMSP00010022171 | 97.1264 | 94.6839 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 91.3669 | 87.7698 | cavia_porcellus | ENSCPOP00000028559 | 95.2024 | 91.4543 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.9856 | 95.2518 | felis_catus | ENSFCAP00000006670 | 96.5957 | 93.9007 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 86.3309 | 83.4532 | ursus_americanus | ENSUAMP00000018376 | 94.9367 | 91.7721 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 85.1799 | 78.2734 | monodelphis_domestica | ENSMODP00000039694 | 87.3156 | 80.236 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 96.6907 | 93.3813 | bison_bison_bison | ENSBBBP00000012194 | 96.1373 | 92.8469 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.8417 | 95.3957 | monodon_monoceros | ENSMMNP00015022556 | 97.7011 | 95.2586 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 96.5468 | 92.9496 | sus_scrofa | ENSSSCP00000018843 | 95.1773 | 91.6312 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.2734 | 95.9712 | microcebus_murinus | ENSMICP00000014925 | 98.557 | 96.2482 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.1223 | 94.1007 | octodon_degus | ENSODEP00000013077 | 97.2622 | 94.2363 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 85.3237 | 80.4317 | jaculus_jaculus | ENSJJAP00000023072 | 93.3858 | 88.0315 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 92.2302 | 88.0576 | jaculus_jaculus | ENSJJAP00000020066 | 96.102 | 91.7541 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 89.9281 | 86.7626 | ovis_aries_rambouillet | ENSOARP00020019439 | 95.1294 | 91.7808 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.4173 | 95.1079 | ovis_aries_rambouillet | ENSOARP00020023106 | 98.4173 | 95.1079 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 92.8058 | 90.0719 | ursus_maritimus | ENSUMAP00000014616 | 97.2851 | 94.4193 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.2662 | 94.8201 | ochotona_princeps | ENSOPRP00000010558 | 97.2662 | 94.8201 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.8417 | 95.3957 | delphinapterus_leucas | ENSDLEP00000009711 | 97.7011 | 95.2586 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.2662 | 94.5324 | physeter_catodon | ENSPCTP00005011682 | 97.1264 | 94.3966 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.1223 | 93.5252 | mus_caroli | MGP_CAROLIEiJ_P0028438 | 97.6845 | 94.0666 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 95.3957 | 91.3669 | mus_musculus | ENSMUSP00000137922 | 93.3803 | 89.4366 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 95.2518 | 92.0863 | dipodomys_ordii | ENSDORP00000027220 | 97.6401 | 94.3953 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 84.0288 | 79.1367 | mustela_putorius_furo | ENSMPUP00000013897 | 86.9048 | 81.8452 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.705 | 97.8417 | aotus_nancymaae | ENSANAP00000040849 | 98.5632 | 97.7011 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 78.2734 | 76.9784 | saimiri_boliviensis_boliviensis | ENSSBOP00000029623 | 96.7971 | 95.1957 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.705 | 97.8417 | saimiri_boliviensis_boliviensis | ENSSBOP00000040150 | 98.705 | 97.8417 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 41.5827 | 38.4173 | vicugna_pacos | ENSVPAP00000004961 | 48.5714 | 44.874 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.5683 | 99.1367 | chlorocebus_sabaeus | ENSCSAP00000003287 | 99.5683 | 99.1367 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 83.5971 | 80.7194 | tupaia_belangeri | ENSTBEP00000007250 | 96.6722 | 93.3444 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 83.5971 | 80 | mesocricetus_auratus | ENSMAUP00000004411 | 97.1572 | 92.9766 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 95.5396 | 92.3741 | phocoena_sinus | ENSPSNP00000019521 | 88.5333 | 85.6 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.554 | 94.6763 | camelus_dromedarius | ENSCDRP00005014449 | 97.554 | 94.6763 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.1295 | 96.6907 | carlito_syrichta | ENSTSYP00000022687 | 98.6975 | 97.2504 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 85.1799 | 82.5899 | panthera_tigris_altaica | ENSPTIP00000019336 | 85.1799 | 82.5899 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.4101 | 94.1007 | rattus_norvegicus | ENSRNOP00000080551 | 97.974 | 94.6454 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.5611 | 95.8273 | prolemur_simus | ENSPSMP00000036601 | 98.7032 | 95.9654 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 96.8345 | 93.6691 | microtus_ochrogaster | ENSMOCP00000026196 | 96.9741 | 93.804 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 58.2734 | 55.1079 | sorex_araneus | ENSSARP00000006540 | 59.2972 | 56.0761 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.4101 | 94.2446 | peromyscus_maniculatus_bairdii | ENSPEMP00000010795 | 97.974 | 94.7902 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 87.3381 | 83.8849 | bos_mutus | ENSBMUP00000011538 | 87.3381 | 83.8849 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 90.0719 | 86.3309 | mus_spicilegus | ENSMSIP00000020995 | 96.7542 | 92.7357 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 86.0432 | 81.7266 | mus_spicilegus | ENSMSIP00000013053 | 93.5837 | 88.8889 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 96.5468 | 93.5252 | cricetulus_griseus_chok1gshd | ENSCGRP00001020239 | 95.8571 | 92.8571 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.6978 | 94.964 | pteropus_vampyrus | ENSPVAP00000011924 | 97.6978 | 94.964 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.1223 | 94.1007 | myotis_lucifugus | ENSMLUP00000015203 | 96.9828 | 93.9655 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 90.3597 | 83.741 | vombatus_ursinus | ENSVURP00010012385 | 86.6207 | 80.2759 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.6978 | 94.964 | rhinolophus_ferrumequinum | ENSRFEP00010018931 | 97.9798 | 95.2381 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.2734 | 95.2518 | neovison_vison | ENSNVIP00000020263 | 97.9914 | 94.9785 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 95.9712 | 91.7986 | bos_taurus | ENSBTAP00000044207 | 96.2482 | 92.0635 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.1223 | 94.5324 | heterocephalus_glaber_female | ENSHGLP00000037759 | 97.2622 | 94.6686 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.5683 | 99.1367 | rhinopithecus_bieti | ENSRBIP00000017243 | 99.4253 | 98.9943 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 96.6907 | 93.0935 | canis_lupus_dingo | ENSCAFP00020014361 | 92.6897 | 89.2414 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.9928 | 96.8345 | sciurus_vulgaris | ENSSVLP00005021952 | 98.9928 | 96.8345 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 90.5036 | 83.8849 | phascolarctos_cinereus | ENSPCIP00000019524 | 85.5782 | 79.3197 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 81.295 | 78.4173 | procavia_capensis | ENSPCAP00000005881 | 81.295 | 78.4173 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 75.2518 | 70.6475 | nannospalax_galili | ENSNGAP00000019000 | 88.4941 | 83.0795 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.1223 | 94.2446 | nannospalax_galili | ENSNGAP00000016090 | 96.7049 | 93.8395 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 88.4892 | 84.3165 | bos_grunniens | ENSBGRP00000031048 | 94.7612 | 90.2928 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.5611 | 95.2518 | bos_grunniens | ENSBGRP00000034393 | 98.2783 | 94.9785 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.6978 | 94.964 | chinchilla_lanigera | ENSCLAP00000011159 | 97.8386 | 95.1009 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.9856 | 95.3957 | cervus_hanglu_yarkandensis | ENSCHYP00000023540 | 97.8448 | 95.2586 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.6978 | 94.5324 | catagonus_wagneri | ENSCWAP00000004259 | 96.7236 | 93.5897 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 83.8849 | 81.1511 | ictidomys_tridecemlineatus | ENSSTOP00000029213 | 92.8344 | 89.8089 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.4173 | 95.9712 | ictidomys_tridecemlineatus | ENSSTOP00000025059 | 98.2759 | 95.8333 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.6978 | 95.1079 | otolemur_garnettii | ENSOGAP00000006618 | 97.4175 | 94.835 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.1367 | 98.4173 | cebus_imitator | ENSCCAP00000013633 | 99.1367 | 98.4173 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 83.1655 | 81.4389 | urocitellus_parryii | ENSUPAP00010024117 | 83.7681 | 82.029 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.8417 | 94.6763 | moschus_moschiferus | ENSMMSP00000013926 | 97.7011 | 94.5402 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 99.5683 | 99.1367 | rhinopithecus_roxellana | ENSRROP00000037251 | 99.4253 | 98.9943 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 97.2662 | 93.813 | mus_pahari | MGP_PahariEiJ_P0035177 | 97.4063 | 93.9481 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 98.8489 | 96.5468 | propithecus_coquereli | ENSPCOP00000011518 | 98.2833 | 95.9943 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 60.8633 | 46.9065 | callorhinchus_milii | ENSCMIP00000029109 | 64.2857 | 49.5441 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 70.2158 | 60.1439 | latimeria_chalumnae | ENSLACP00000014588 | 75.7764 | 64.9068 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 65.4676 | 53.5252 | xenopus_tropicalis | ENSXETP00000074586 | 68.9394 | 56.3636 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 84.6043 | 77.1223 | chrysemys_picta_bellii | ENSCPBP00000028477 | 87.2404 | 79.5252 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 69.9281 | 61.8705 | sphenodon_punctatus | ENSSPUP00000006624 | 82.5127 | 73.0051 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 74.1007 | 66.187 | crocodylus_porosus | ENSCPRP00005020569 | 81.876 | 73.132 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 60.7194 | 49.2086 | laticauda_laticaudata | ENSLLTP00000005379 | 66.248 | 53.6892 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 59.1367 | 47.1942 | notechis_scutatus | ENSNSUP00000002159 | 68.2724 | 54.4851 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 60 | 48.2014 | pseudonaja_textilis | ENSPTXP00000023650 | 61.9614 | 49.7771 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 74.2446 | 64.3165 | gallus_gallus | ENSGALP00010044501 | 78.0635 | 67.6248 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 71.9424 | 62.5899 | meleagris_gallopavo | ENSMGAP00000005883 | 73.5294 | 63.9706 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 73.813 | 65.8993 | serinus_canaria | ENSSCAP00000012151 | 83.1442 | 74.2301 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 82.0144 | 73.6691 | parus_major | ENSPMJP00000026965 | 86.4947 | 77.6935 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 67.3381 | 56.4029 | anolis_carolinensis | ENSACAP00000012636 | 66.9528 | 56.0801 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 62.8777 | 51.3669 | naja_naja | ENSNNAP00000003067 | 68.3881 | 55.8685 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 56.8345 | 47.6259 | podarcis_muralis | ENSPMRP00000034380 | 60.6759 | 50.8449 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 70.3597 | 63.3094 | geospiza_fortis | ENSGFOP00000012697 | 84.0206 | 75.6014 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 82.5899 | 73.9568 | taeniopygia_guttata | ENSTGUP00000021767 | 86.4458 | 77.4096 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 58.4173 | 49.7842 | salvator_merianae | ENSSMRP00000000368 | 70.7317 | 60.2787 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 64.8921 | 58.9928 | pelodiscus_sinensis | ENSPSIP00000004663 | 85.4167 | 77.6515 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 83.0216 | 75.3957 | gopherus_evgoodei | ENSGEVP00005008685 | 86.1194 | 78.209 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 80 | 73.2374 | chelonoidis_abingdonii | ENSCABP00000011157 | 85.1455 | 77.9479 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 77.1223 | 69.2086 | struthio_camelus_australis | ENSSCUP00000019005 | 84.2767 | 75.6289 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 81.295 | 73.0935 | strigops_habroptila | ENSSHBP00005018662 | 85.8663 | 77.2036 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 84.7482 | 77.554 | terrapene_carolina_triunguis | ENSTMTP00000011084 | 86.7452 | 79.3814 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 82.446 | 74.3885 | aquila_chrysaetos_chrysaetos | ENSACCP00020021081 | 86.9499 | 78.4522 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 74.2446 | 65.8993 | ficedula_albicollis | ENSFALP00000006537 | 82.1656 | 72.9299 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 75.9712 | 64.7482 | coturnix_japonica | ENSCJPP00005014615 | 77.6471 | 66.1765 |
| ENSG00000108256 | homo_sapiens | ENSP00000225388 | 64.3165 | 51.223 | leptobrachium_leishanense | ENSLLEP00000025370 | 67.9331 | 54.1033 |
| Go ID |
Go term |
No. evidence |
Entries |
Species |
Category |
| GO:0003723 | enables RNA binding | 3 | HDA,IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | is_active_in nucleoplasm | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0010494 | is_active_in cytoplasmic stress granule | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0016020 | located_in membrane | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0016604 | located_in nuclear body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0042788 | is_active_in polysomal ribosome | 2 | IBA,IDA | Homo_sapiens(9606) | Component |