RBP Type
Non-canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
E3 ligase
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000108094 Gene ID
8453 Accession
2552
Symbol
CUL2
Alias
NA
Full Name
cullin 2
Status
Confidence
Length
118503 bases
Strand
Minus strand
Position
10 : 35008504 - 35127006
RNA binding domain
N.A.
Summary
Enables ubiquitin protein ligase binding activity. Predicted to be involved in SCF-dependent proteasomal ubiquitin-dependent protein catabolic process and protein ubiquitination. Predicted to act upstream of or within protein catabolic process. Located in nucleoplasm. Part of Cul2-RING ubiquitin ligase complex. [provided by Alliance of Genome Resources, Apr 2022]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
35189958 circCUL2 induces an inflammatory CAF phenotype in pancreatic ductal adenocarcinoma via the activation of the MyD88-dependent NF-κB signaling pathway.
Shangyou Zheng
2022-02-21
Journal of experimental & clinical cancer research : CR
Name
Transcript ID
bp
Protein
Translation ID
CUL2-210
ENST00000626172
4116
706aa
ENSP00000485986
CUL2-201
ENST00000374746
3867
694aa
ENSP00000363878
CUL2-209
ENST00000537177
4223
758aa
ENSP00000444856
CUL2-236
ENST00000690123
3984
682aa
ENSP00000508924
CUL2-203
ENST00000374749
4183
745aa
ENSP00000363881
CUL2-202
ENST00000374748
4312
745aa
ENSP00000363880
CUL2-204
ENST00000374751
4233
745aa
ENSP00000363883
CUL2-246
ENST00000691974
3187
745aa
ENSP00000508908
CUL2-229
ENST00000688571
3026
758aa
ENSP00000509574
CUL2-237
ENST00000690393
2934
745aa
ENSP00000510691
CUL2-235
ENST00000690092
2599
711aa
ENSP00000508675
CUL2-205
ENST00000374754
2828
90aa
ENSP00000363886
CUL2-226
ENST00000688252
4690
No protein
-
CUL2-241
ENST00000691272
2739
240aa
ENSP00000508531
CUL2-223
ENST00000687282
3567
No protein
-
CUL2-227
ENST00000688390
3575
No protein
-
CUL2-239
ENST00000691201
2663
716aa
ENSP00000509778
CUL2-232
ENST00000689036
2204
No protein
-
CUL2-222
ENST00000687194
2818
242aa
ENSP00000509491
CUL2-225
ENST00000688044
2871
No protein
-
CUL2-240
ENST00000691263
2717
742aa
ENSP00000510240
CUL2-244
ENST00000691858
2750
240aa
ENSP00000509932
CUL2-245
ENST00000691962
2663
394aa
ENSP00000508699
CUL2-230
ENST00000688705
2079
No protein
-
CUL2-247
ENST00000692456
2515
No protein
-
CUL2-211
ENST00000673636
2839
745aa
ENSP00000501215
CUL2-224
ENST00000687524
2738
753aa
ENSP00000510171
CUL2-249
ENST00000693739
2691
242aa
ENSP00000508657
CUL2-248
ENST00000692866
3623
No protein
-
CUL2-219
ENST00000686302
1707
472aa
ENSP00000510342
CUL2-216
ENST00000685681
2792
166aa
ENSP00000508572
CUL2-206
ENST00000421317
2646
764aa
ENSP00000414095
CUL2-213
ENST00000684965
2238
256aa
ENSP00000509100
CUL2-242
ENST00000691457
3182
141aa
ENSP00000509618
CUL2-228
ENST00000688541
2183
242aa
ENSP00000509171
CUL2-234
ENST00000689705
643
101aa
ENSP00000508869
CUL2-233
ENST00000689330
1734
249aa
ENSP00000509501
CUL2-218
ENST00000686271
582
No protein
-
CUL2-220
ENST00000686748
895
148aa
ENSP00000509655
CUL2-238
ENST00000690789
883
112aa
ENSP00000510162
CUL2-215
ENST00000685488
1073
49aa
ENSP00000510204
CUL2-243
ENST00000691635
796
238aa
ENSP00000509428
CUL2-221
ENST00000686903
1147
249aa
ENSP00000508936
CUL2-212
ENST00000684955
777
87aa
ENSP00000508946
CUL2-207
ENST00000468804
712
141aa
ENSP00000509962
CUL2-214
ENST00000685421
699
204aa
ENSP00000509605
CUL2-217
ENST00000686156
847
204aa
ENSP00000509166
CUL2-231
ENST00000688736
948
204aa
ENSP00000510643
CUL2-208
ENST00000478044
483
78aa
ENSP00000509576
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
13328 Parkinson Disease 3.32E-5 19915575 13349 Parkinson Disease 1.48E-5 19915575 13350 Parkinson Disease 2.68E-5 19915575 13358 Parkinson Disease 2.13E-5 19915575 13365 Parkinson Disease 2.2E-5 19915575 13366 Parkinson Disease 1.43E-5 19915575 13374 Parkinson Disease 2.14E-5 19915575 13375 Parkinson Disease 2.31E-5 19915575
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
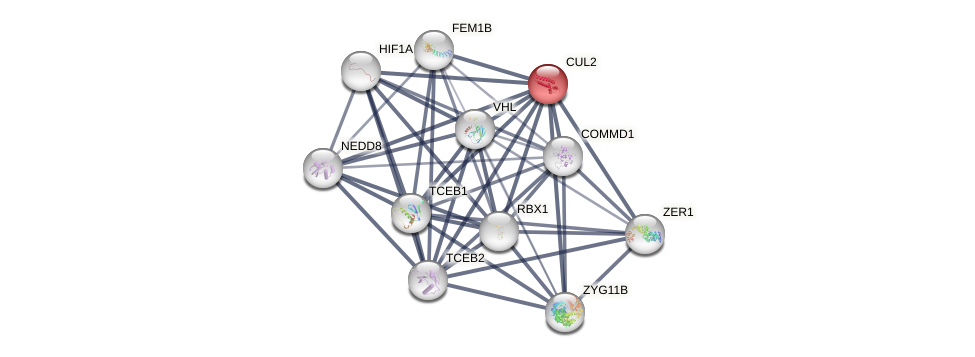
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000082 involved_in G1/S transition of mitotic cell cycle 1 TAS Homo_sapiens(9606) Process GO:0004842 enables ubiquitin-protein transferase activity 1 TAS Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005654 located_in nucleoplasm 2 IDA,TAS Homo_sapiens(9606) Component GO:0005730 located_in nucleolus 1 IEA Homo_sapiens(9606) Component GO:0005829 located_in cytosol 1 TAS Homo_sapiens(9606) Component GO:0016567 involved_in protein ubiquitination 3 IBA,IEA,TAS Homo_sapiens(9606) Process GO:0019005 part_of SCF ubiquitin ligase complex 1 IBA Homo_sapiens(9606) Component GO:0030674 enables protein-macromolecule adaptor activity 1 IBA Homo_sapiens(9606) Function GO:0030891 part_of VCB complex 1 IEA Homo_sapiens(9606) Component GO:0031146 involved_in SCF-dependent proteasomal ubiquitin-dependent protein catabolic process 1 IBA Homo_sapiens(9606) Process GO:0031462 part_of Cul2-RING ubiquitin ligase complex 2 IBA,IDA Homo_sapiens(9606) Component GO:0031625 enables ubiquitin protein ligase binding 2 IBA,IDA Homo_sapiens(9606) Function GO:0044877 enables protein-containing complex binding 1 IEA Homo_sapiens(9606) Function GO:0097193 involved_in intrinsic apoptotic signaling pathway 1 TAS Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.