| RBP Type |
Non-canonical_RBPs |
| Diseases |
N.A. |
| Drug |
N.A. |
| Main interacting RNAs |
mRNA |
| Moonlighting functions |
N.A. |
| Localizations |
N.A. |
|
BulkPerturb-seq
|
N.A. |
| Ensembl ID | ENSG00000105298 | Gene ID |
58509 |
Accession |
29938 |
| Symbol |
CACTIN |
Alias |
|
Full Name |
|
| Status |
Confidence |
Length |
16171 bases |
Strand |
Minus strand |
| Position |
19 : 3610645 - 3626815 |
RNA binding domain |
N.A. |
| Summary |
|
RNA binding domains (RBDs)

| Protein |
Domain |
Pfam ID |
E-value |
Domain number |
RNA binding proteomes (RBPomes)
| Pubmed ID |
Full Name |
Cell |
Author |
Time |
Doi |
Literatures on RNA binding capacity
| Pubmed ID |
Title |
Author |
Time |
Journal |
| Name |
Transcript ID |
bp |
Protein |
Translation ID |
| CACTIN-208 |
ENST00000591726 |
699 |
117aa |
ENSP00000464898 |
| CACTIN-210 |
ENST00000592721 |
1220 |
291aa |
ENSP00000467149 |
| CACTIN-203 |
ENST00000429344 |
3583 |
758aa |
ENSP00000415078 |
| CACTIN-204 |
ENST00000585942 |
2810 |
758aa |
ENSP00000465751 |
| CACTIN-202 |
ENST00000248420 |
2671 |
758aa |
ENSP00000248420 |
| CACTIN-201 |
ENST00000221899 |
2797 |
758aa |
ENSP00000221899 |
| CACTIN-207 |
ENST00000589321 |
1708 |
141aa |
ENSP00000466600 |
| CACTIN-205 |
ENST00000587175 |
2216 |
No protein |
- |
| CACTIN-206 |
ENST00000588749 |
391 |
130aa |
ENSP00000467788 |
| CACTIN-209 |
ENST00000591978 |
2283 |
No protein |
- |
| Pathway ID |
Pathway Name |
Source |
| | |
| ensgID |
Trait |
pValue |
Pubmed ID |
| 70495 | Insulin-Like Growth Factor Binding Protein 3 | 6E-6 | 23251661 |
| ensgID | SNP | Chromosome | Position |
Trait | PubmedID | Or or BEAT |
EFO ID |
|---|
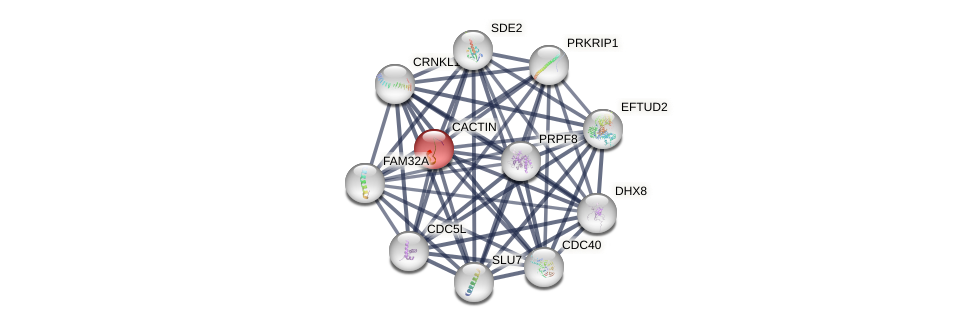
Protein-Protein Interaction (PPI)
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSG00000105298 | homo_sapiens | ENSP00000268485 | 30.6733 | 17.9551 | homo_sapiens | ENSP00000415078 | 16.2269 | 9.49868 |
| ENSG00000105298 | homo_sapiens | ENSP00000391759 | 30.566 | 17.3585 | homo_sapiens | ENSP00000415078 | 10.686 | 6.0686 |
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 73.3509 | 72.2955 | nomascus_leucogenys | ENSNLEP00000003096 | 88.96 | 87.68 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 93.5356 | 93.2718 | pan_paniscus | ENSPPAP00000009948 | 99.8592 | 99.5775 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 99.3404 | 98.8127 | pongo_abelii | ENSPPYP00000010523 | 99.0789 | 98.5526 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 99.4723 | 99.2084 | pan_troglodytes | ENSPTRP00000017458 | 99.7355 | 99.4709 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 99.8681 | 99.4723 | gorilla_gorilla | ENSGGOP00000012221 | 99.7365 | 99.3412 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 87.467 | 82.058 | dasypus_novemcinctus | ENSDNOP00000028519 | 88.1649 | 82.7128 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 43.9314 | 42.2164 | echinops_telfairi | ENSETEP00000001009 | 52.6899 | 50.6329 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 98.4169 | 97.3615 | callithrix_jacchus | ENSCJAP00000027515 | 97.5163 | 96.4706 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 98.4169 | 97.8892 | cercocebus_atys | ENSCATP00000039393 | 98.1579 | 97.6316 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 98.6807 | 97.8892 | macaca_mulatta | ENSMMUP00000031971 | 98.4211 | 97.6316 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 93.4037 | 92.3483 | macaca_nemestrina | ENSMNEP00000029570 | 95.0336 | 93.9597 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 98.6807 | 98.0211 | papio_anubis | ENSPANP00000028354 | 98.4211 | 97.7632 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 92.7441 | 91.5567 | mandrillus_leucophaeus | ENSMLEP00000003938 | 94.8718 | 93.6572 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 77.0449 | 75.7256 | tursiops_truncatus | ENSTTRP00000005452 | 78.2842 | 76.9437 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.4591 | 92.6121 | vulpes_vulpes | ENSVVUP00000004272 | 95.2128 | 93.3511 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 91.4248 | 86.6755 | mus_spretus | MGP_SPRETEiJ_P0025288 | 90.117 | 85.4356 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.723 | 91.9525 | equus_asinus | ENSEASP00005053090 | 95.3519 | 92.5631 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 89.314 | 87.467 | capra_hircus | ENSCHIP00000033031 | 95.6215 | 93.6441 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 21.8997 | 20.9763 | loxodonta_africana | ENSLAFP00000024587 | 92.7374 | 88.8268 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 93.6675 | 91.029 | panthera_leo | ENSPLOP00000024337 | 94.2895 | 91.6335 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 90.7652 | 89.0501 | ailuropoda_melanoleuca | ENSAMEP00000040607 | 89.1192 | 87.4352 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.3272 | 92.3483 | bos_indicus_hybrid | ENSBIXP00005034171 | 94.5767 | 92.5926 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.8549 | 91.9525 | equus_caballus | ENSECAP00000038445 | 95.4847 | 92.5631 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.4591 | 92.6121 | canis_lupus_familiaris | ENSCAFP00845012109 | 95.2128 | 93.3511 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 79.9472 | 77.3087 | panthera_pardus | ENSPPRP00000013950 | 93.3744 | 90.2928 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 87.7309 | 85.0923 | marmota_marmota_marmota | ENSMMMP00000019026 | 92.233 | 89.4591 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 93.9314 | 90.7652 | balaenoptera_musculus | ENSBMSP00010011225 | 94.3046 | 91.1258 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 93.2718 | 90.6332 | cavia_porcellus | ENSCPOP00000003502 | 93.6424 | 90.9934 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 93.6675 | 91.2929 | felis_catus | ENSFCAP00000021528 | 94.1645 | 91.7772 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.723 | 92.876 | ursus_americanus | ENSUAMP00000021868 | 95.4787 | 93.617 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.591 | 92.4802 | bison_bison_bison | ENSBBBP00000027067 | 95.2191 | 93.0943 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.3272 | 91.029 | monodon_monoceros | ENSMMNP00015001017 | 94.702 | 91.3907 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 95.1187 | 92.0844 | sus_scrofa | ENSSSCP00000056347 | 95.6233 | 92.5729 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.723 | 93.1398 | microcebus_murinus | ENSMICP00000007560 | 95.3519 | 93.7583 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 82.8496 | 78.1003 | octodon_degus | ENSODEP00000020895 | 86.5014 | 81.5427 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.723 | 91.9525 | octodon_degus | ENSODEP00000019962 | 94.9735 | 92.1958 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 91.5567 | 87.5989 | jaculus_jaculus | ENSJJAP00000020301 | 92.7807 | 88.7701 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.8549 | 92.7441 | ovis_aries_rambouillet | ENSOARP00020023863 | 95.1058 | 92.9894 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 13.5884 | 11.0818 | ursus_maritimus | ENSUMAP00000009770 | 54.4974 | 44.4444 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.1953 | 90.7652 | delphinapterus_leucas | ENSDLEP00000021195 | 94.5695 | 91.1258 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 93.5356 | 90.6332 | physeter_catodon | ENSPCTP00005025952 | 94.1567 | 91.2351 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 12.2691 | 10.8179 | mus_caroli | MGP_CAROLIEiJ_P0084852 | 71.5385 | 63.0769 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 91.8206 | 87.2032 | mus_caroli | MGP_CAROLIEiJ_P0024114 | 91.0995 | 86.5183 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 91.5567 | 86.8074 | mus_musculus | ENSMUSP00000059533 | 89.8964 | 85.2332 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 54.2216 | 52.7704 | dipodomys_ordii | ENSDORP00000003825 | 91.9463 | 89.4855 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.591 | 92.0844 | mustela_putorius_furo | ENSMPUP00000004002 | 95.3457 | 92.8191 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 98.285 | 96.8338 | aotus_nancymaae | ENSANAP00000034093 | 94.1846 | 92.7939 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 76.3852 | 75.066 | saimiri_boliviensis_boliviensis | ENSSBOP00000035864 | 98.3022 | 96.6044 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 42.4802 | 40.5013 | vicugna_pacos | ENSVPAP00000003296 | 46.8705 | 44.687 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 89.5778 | 89.1821 | chlorocebus_sabaeus | ENSCSAP00000007329 | 98.6919 | 98.2558 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 89.7098 | 85.3562 | mesocricetus_auratus | ENSMAUP00000010922 | 85.1064 | 80.9762 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.3272 | 90.7652 | phocoena_sinus | ENSPSNP00000026353 | 94.702 | 91.1258 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 95.1187 | 93.0079 | camelus_dromedarius | ENSCDRP00005008353 | 95.8777 | 93.75 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 90.5013 | 86.2797 | rattus_norvegicus | ENSRNOP00000027926 | 89.7906 | 85.6021 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.723 | 93.0079 | prolemur_simus | ENSPSMP00000027363 | 95.0993 | 93.3775 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 91.029 | 87.3351 | microtus_ochrogaster | ENSMOCP00000011745 | 92.4933 | 88.7399 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 49.2084 | 41.8206 | sorex_araneus | ENSSARP00000008335 | 73.57 | 62.5247 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 85.0923 | 81.5303 | peromyscus_maniculatus_bairdii | ENSPEMP00000025250 | 94.2982 | 90.3509 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.4591 | 92.3483 | bos_mutus | ENSBMUP00000024121 | 94.709 | 92.5926 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 91.4248 | 86.6755 | mus_spicilegus | ENSMSIP00000028624 | 90 | 85.3247 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 91.8206 | 87.8628 | cricetulus_griseus_chok1gshd | ENSCGRP00001022888 | 92.5532 | 88.5638 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 85.3562 | 83.905 | pteropus_vampyrus | ENSPVAP00000004075 | 77.9518 | 76.6265 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 92.7441 | 90.3694 | myotis_lucifugus | ENSMLUP00000003214 | 92.9894 | 90.6085 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 93.2718 | 90.8971 | rhinolophus_ferrumequinum | ENSRFEP00010031027 | 93.7666 | 91.3793 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.591 | 92.3483 | neovison_vison | ENSNVIP00000008309 | 95.3457 | 93.0851 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.3272 | 92.2164 | bos_taurus | ENSBTAP00000013697 | 94.5767 | 92.4603 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 93.0079 | 91.1609 | heterocephalus_glaber_female | ENSHGLP00000043519 | 89.3536 | 87.5792 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 99.0765 | 98.4169 | rhinopithecus_bieti | ENSRBIP00000001486 | 93.408 | 92.7861 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.3272 | 92.4802 | canis_lupus_dingo | ENSCAFP00020023201 | 95.0798 | 93.2181 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.8549 | 92.876 | sciurus_vulgaris | ENSSVLP00005030207 | 95.4847 | 93.4927 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 79.2876 | 77.1768 | procavia_capensis | ENSPCAP00000010376 | 75.125 | 73.125 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 90.5013 | 86.5435 | nannospalax_galili | ENSNGAP00000021573 | 90.1445 | 86.2024 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.4591 | 92.3483 | bos_grunniens | ENSBGRP00000033350 | 94.709 | 92.5926 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 92.7441 | 90.5013 | chinchilla_lanigera | ENSCLAP00000015586 | 92.9894 | 90.7407 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.591 | 92.7441 | cervus_hanglu_yarkandensis | ENSCHYP00000005556 | 91.8054 | 90.0128 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.9868 | 91.8206 | catagonus_wagneri | ENSCWAP00000014134 | 95.4907 | 92.3077 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 91.029 | 89.0501 | ictidomys_tridecemlineatus | ENSSTOP00000001110 | 95.8333 | 93.75 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 85.752 | 84.4327 | otolemur_garnettii | ENSOGAP00000009297 | 96.0118 | 94.5347 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 98.6807 | 97.3615 | cebus_imitator | ENSCCAP00000021798 | 98.2917 | 96.9777 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 93.9314 | 91.9525 | urocitellus_parryii | ENSUPAP00010010551 | 94.8069 | 92.8096 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.0633 | 92.0844 | moschus_moschiferus | ENSMMSP00000031067 | 94.8138 | 92.8191 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 91.8206 | 90.7652 | rhinopithecus_roxellana | ENSRROP00000016506 | 96.1326 | 95.0276 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 91.2929 | 86.4116 | mus_pahari | MGP_PahariEiJ_P0087263 | 92.0213 | 87.1011 |
| ENSG00000105298 | homo_sapiens | ENSP00000415078 | 94.9868 | 93.2718 | propithecus_coquereli | ENSPCOP00000009256 | 95.2381 | 93.5185 |
| Go ID |
Go term |
No. evidence |
Entries |
Species |
Category |
| GO:0000398 | involved_in mRNA splicing, via spliceosome | 2 | IC,IMP | Homo_sapiens(9606) | Process |
| GO:0001933 | involved_in negative regulation of protein phosphorylation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005681 | part_of spliceosomal complex | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0016607 | located_in nuclear speck | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0031665 | involved_in negative regulation of lipopolysaccharide-mediated signaling pathway | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032088 | involved_in negative regulation of NF-kappaB transcription factor activity | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032688 | involved_in negative regulation of interferon-beta production | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032717 | involved_in negative regulation of interleukin-8 production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032720 | involved_in negative regulation of tumor necrosis factor production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0034122 | involved_in negative regulation of toll-like receptor signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0043124 | involved_in negative regulation of I-kappaB kinase/NF-kappaB signaling | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045087 | involved_in innate immune response | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045292 | involved_in mRNA cis splicing, via spliceosome | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0045824 | involved_in negative regulation of innate immune response | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0060339 | involved_in negative regulation of type I interferon-mediated signaling pathway | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0071013 | part_of catalytic step 2 spliceosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0071222 | involved_in cellular response to lipopolysaccharide | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071347 | involved_in cellular response to interleukin-1 | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071356 | involved_in cellular response to tumor necrosis factor | 1 | IDA | Homo_sapiens(9606) | Process |