Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000101916 | Gene ID | 51311 | Accession | 15632 |
| Symbol | TLR8 | Alias | CD288;IMD98;TLR-8;hTLR8 | Full Name | toll like receptor 8 |
| Status | Confidence | Length | 16550 bases | Strand | Plus strand |
| Position | X : 12906620 - 12923169 | RNA binding domain | N.A. | ||
| Summary | The protein encoded by this gene is a member of the Toll-like receptor (TLR) family which plays a fundamental role in pathogen recognition and activation of innate immunity. TLRs are highly conserved from Drosophila to humans and share structural and functional similarities. They recognize pathogen-associated molecular patterns (PAMPs) that are expressed on infectious agents, and mediate the production of cytokines necessary for the development of effective immunity. The various TLRs exhibit different patterns of expression. This gene is predominantly expressed in lung and peripheral blood leukocytes, and lies in close proximity to another family member, TLR7, on chromosome X. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 34677064 | Immunogenicity of In Vitro-Transcribed RNA. | Xin Mu | 2021-11-02 | Accounts of chemical research |
| 37513775 | The Potential of Probiotics as Ingestible Adjuvants and Immune Modulators for Antiviral Immunity and Management of SARS-CoV-2 Infection and COVID-19. | Sophie Tomkinson | 2023-07-11 | Pathogens (Basel, Switzerland) |
| 30699960 | RNA Modifications Modulate Activation of Innate Toll-Like Receptors. | Isabel Freund | 2019-01-29 | Genes |
| 32933031 | Neutrophil Extracellular Traps and By-Products Play a Key Role in COVID-19: Pathogenesis, Risk Factors, and Therapy. | R Alain Thierry | 2020-09-11 | Journal of clinical medicine |
| 27988432 | TLRs, future potential therapeutic targets for RA. | A Hatem Elshabrawy | 2017-02-01 | Autoimmunity reviews |
| 25431922 | Looking into a conceptual framework of ROS-miRNA-atrial fibrillation. | Seahyoung Lee | 2014-11-26 | International journal of molecular sciences |
| 24062747 | Aberrant Type I Interferon Regulation in Autoimmunity: Opposite Directions in MS and SLE, Shaped by Evolution and Body Ecology. | T Anthony Reder | 2013-09-17 | Frontiers in immunology |
| 27455396 | Discriminating self from non-self in nucleic acid sensing. | Martin Schlee | 2016-09-01 | Nature reviews. Immunology |
| 23965954 | Principles of miRNA-target regulation in metazoan models. | Epaminondas Doxakis | 2013-08-07 | International journal of molecular sciences |
| 27769507 | Endogenous Retroelements and the Host Innate Immune Sensors. | X Mu | 2016-01-01 | Advances in immunology |
| 36986379 | IFN-Induced PARPs-Sensors of Foreign Nucleic Acids? | Katharina Biaesch | 2023-03-14 | Pathogens (Basel, Switzerland) |
| 28987220 | Viral infections in allergy and immunology: How allergic inflammation influences viral infections and illness. | R Michael Edwards | 2017-10-01 | The Journal of allergy and clinical immunology |
| 33650968 | In vivo reconstitution finds multivalent RNA-RNA interactions as drivers of mesh-like condensates. | Weirui Ma | 2021-03-02 | eLife |
| 28530246 | Short single-stranded DNA degradation products augment the activation of Toll-like receptor 9. | Jelka Pohar | 2017-05-22 | Nature communications |
| 34414672 | Phase 1 study in healthy participants of the safety, pharmacokinetics, and pharmacodynamics of enpatoran (M5049), a dual antagonist of toll-like receptors 7 and 8. | Andreas Port | 2021-10-01 | Pharmacology research & perspectives |
| 36267192 | mRNA vaccines: A novel weapon to control infectious diseases. | Yuying Tian | 2022-01-01 | Frontiers in microbiology |
| 31778653 | TLR8 Is a Sensor of RNase T2 Degradation Products. | Wilhelm Greulich | 2019-11-27 | Cell |
| 25487859 | Base modification strategies to modulate immune stimulation by an siRNA. | P Anne Rachel Valenzuela | 2015-01-19 | Chembiochem : a European journal of chemical biology |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| TLR8-201 | ENST00000218032 | 4216 | 1041aa | ENSP00000218032 |
| TLR8-202 | ENST00000311912 | 3367 | 1059aa | ENSP00000312082 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 63725 | Celiac Disease | 6E-8 | 20190752 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
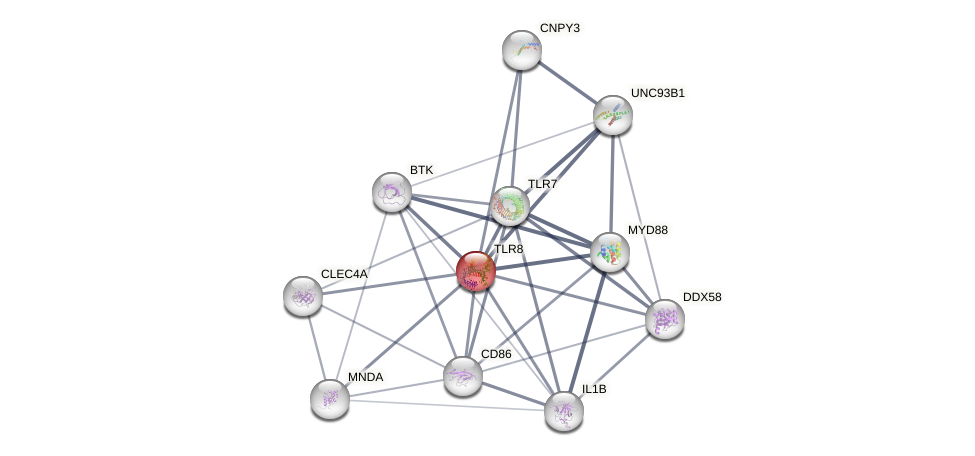
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000101916 | homo_sapiens | ENSP00000370034 | 59.4852 | 42.04 | homo_sapiens | ENSP00000218032 | 59.9424 | 42.3631 |
| ENSG00000101916 | homo_sapiens | ENSP00000306864 | 30.6092 | 18.8707 | homo_sapiens | ENSP00000218032 | 19.7887 | 12.1998 |
| ENSG00000101916 | homo_sapiens | ENSP00000215539 | 39.5041 | 25.6198 | homo_sapiens | ENSP00000218032 | 22.9587 | 14.8895 |
| ENSG00000101916 | homo_sapiens | ENSP00000363089 | 39.8093 | 23.0036 | homo_sapiens | ENSP00000218032 | 32.0845 | 18.5399 |
| ENSG00000101916 | homo_sapiens | ENSP00000344242 | 37.1882 | 21.9955 | homo_sapiens | ENSP00000218032 | 15.7541 | 9.31796 |
| ENSG00000101916 | homo_sapiens | ENSP00000291592 | 41.6342 | 26.07 | homo_sapiens | ENSP00000218032 | 10.2786 | 6.43612 |
| ENSG00000101916 | homo_sapiens | ENSP00000367157 | 37.0909 | 23.6364 | homo_sapiens | ENSP00000218032 | 9.79827 | 6.244 |
| ENSG00000101916 | homo_sapiens | ENSP00000379880 | 38.9961 | 20.4633 | homo_sapiens | ENSP00000218032 | 9.70221 | 5.09126 |
| ENSG00000101916 | homo_sapiens | ENSP00000328625 | 36.9942 | 21.0983 | homo_sapiens | ENSP00000218032 | 24.5917 | 14.025 |
| ENSG00000101916 | homo_sapiens | ENSP00000308925 | 38.1011 | 20.8385 | homo_sapiens | ENSP00000218032 | 29.683 | 16.2344 |
| ENSG00000101916 | homo_sapiens | ENSP00000296795 | 42.0354 | 25.5531 | homo_sapiens | ENSP00000218032 | 36.5034 | 22.1902 |
| ENSG00000101916 | homo_sapiens | ENSP00000496355 | 40.7925 | 23.7762 | homo_sapiens | ENSP00000218032 | 33.6215 | 19.5965 |
| ENSG00000101916 | homo_sapiens | ENSP00000332668 | 34.5609 | 23.2295 | homo_sapiens | ENSP00000218032 | 11.7195 | 7.87704 |
| ENSG00000101916 | homo_sapiens | ENSP00000298386 | 31.8302 | 17.6393 | homo_sapiens | ENSP00000218032 | 23.0548 | 12.7762 |
| ENSG00000101916 | homo_sapiens | ENSP00000494425 | 39.4133 | 23.0867 | homo_sapiens | ENSP00000218032 | 29.683 | 17.3871 |
| ENSG00000101916 | homo_sapiens | ENSP00000260061 | 38.2175 | 24.4713 | homo_sapiens | ENSP00000218032 | 24.3036 | 15.562 |
| ENSG00000101916 | homo_sapiens | ENSP00000354932 | 39.6947 | 21.5013 | homo_sapiens | ENSP00000218032 | 29.9712 | 16.2344 |
| ENSG00000101916 | homo_sapiens | ENSP00000361185 | 32.9091 | 17.4545 | homo_sapiens | ENSP00000218032 | 17.3871 | 9.2219 |
| ENSG00000101916 | homo_sapiens | ENSP00000303248 | 31.4399 | 17.1731 | homo_sapiens | ENSP00000218032 | 22.8626 | 12.488 |
| ENSG00000101916 | homo_sapiens | ENSP00000294635 | 12.911 | 7.69848 | homo_sapiens | ENSP00000218032 | 15.4659 | 9.2219 |
| ENSG00000101916 | homo_sapiens | ENSP00000424718 | 36.5578 | 20.2261 | homo_sapiens | ENSP00000218032 | 27.9539 | 15.4659 |
| ENSG00000101916 | homo_sapiens | ENSP00000256447 | 39.4856 | 23.9032 | homo_sapiens | ENSP00000218032 | 25.072 | 15.1777 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 98.7512 | 98.1748 | nomascus_leucogenys | ENSNLEP00000011109 | 98.7512 | 98.1748 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 99.9039 | 99.7118 | pan_paniscus | ENSPPAP00000033784 | 99.9039 | 99.7118 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 99.3276 | 98.367 | pongo_abelii | ENSPPYP00000022512 | 97.6393 | 96.695 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 99.8079 | 99.5197 | pan_troglodytes | ENSPTRP00000062546 | 98.1114 | 97.8281 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 99.8079 | 99.6158 | gorilla_gorilla | ENSGGOP00000012021 | 98.1114 | 97.9226 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 77.8098 | 63.3045 | ornithorhynchus_anatinus | ENSOANP00000039956 | 77.4379 | 63.0019 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 77.2334 | 64.8415 | sarcophilus_harrisii | ENSSHAP00000016454 | 77.3821 | 64.9663 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 60.8069 | 51.1047 | notamacropus_eugenii | ENSMEUP00000014999 | 61.6358 | 51.8014 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.3026 | 76.561 | choloepus_hoffmanni | ENSCHOP00000007492 | 84.1706 | 75.545 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.6868 | 78.2901 | dasypus_novemcinctus | ENSDNOP00000030741 | 86.0174 | 78.5921 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 81.172 | 70.6052 | erinaceus_europaeus | ENSEEUP00000006108 | 80.9387 | 70.4023 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 64.2651 | 58.4054 | echinops_telfairi | ENSETEP00000001839 | 64.7002 | 58.8008 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 96.0615 | 92.6993 | callithrix_jacchus | ENSCJAP00000007141 | 96.0615 | 92.6993 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 97.9827 | 96.5418 | cercocebus_atys | ENSCATP00000043467 | 96.3173 | 94.9008 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 97.7906 | 96.4457 | macaca_fascicularis | ENSMFAP00000005530 | 98.1678 | 96.8177 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 98.0788 | 96.6378 | macaca_mulatta | ENSMMUP00000018720 | 98.1731 | 96.7308 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 97.5985 | 96.0615 | macaca_nemestrina | ENSMNEP00000003246 | 95.9396 | 94.4287 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 97.8867 | 96.5418 | papio_anubis | ENSPANP00000006798 | 96.4049 | 95.0804 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 97.7906 | 96.2536 | mandrillus_leucophaeus | ENSMLEP00000026501 | 96.1284 | 94.6176 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 84.438 | 76.561 | tursiops_truncatus | ENSTTRP00000003026 | 83.3966 | 75.6167 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.7829 | 77.6177 | vulpes_vulpes | ENSVVUP00000026250 | 86.0308 | 77.842 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 81.6523 | 70.0288 | mus_spretus | MGP_SPRETEiJ_P0097157 | 82.3643 | 70.6395 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 86.2632 | 77.1374 | equus_asinus | ENSEASP00005037366 | 86.2632 | 77.1374 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 83.6695 | 73.9673 | capra_hircus | ENSCHIP00000016328 | 80.2765 | 70.9677 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.3026 | 77.4256 | loxodonta_africana | ENSLAFP00000009826 | 86.2136 | 78.2524 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.975 | 77.2334 | panthera_leo | ENSPLOP00000029301 | 86.1405 | 77.3821 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.3987 | 76.8492 | ailuropoda_melanoleuca | ENSAMEP00000020826 | 85.563 | 76.9971 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 83.7656 | 73.9673 | bos_indicus_hybrid | ENSBIXP00005029629 | 84.4143 | 74.5402 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 68.5879 | 56.9645 | oryctolagus_cuniculus | ENSOCUP00000026782 | 69.3204 | 57.5728 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.975 | 76.8492 | equus_caballus | ENSECAP00000089215 | 86.2235 | 77.0713 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.6868 | 77.2334 | canis_lupus_familiaris | ENSCAFP00845039281 | 85.9345 | 77.4566 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 86.1671 | 77.4256 | panthera_pardus | ENSPPRP00000020697 | 86.0844 | 77.3512 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 77.4256 | 69.7406 | marmota_marmota_marmota | ENSMMMP00000004782 | 86.4807 | 77.897 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 84.2459 | 76.3689 | balaenoptera_musculus | ENSBMSP00010026149 | 84.7343 | 76.8116 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 83.7656 | 74.7358 | cavia_porcellus | ENSCPOP00000017959 | 84.2512 | 75.1691 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.975 | 77.2334 | felis_catus | ENSFCAP00000017550 | 86.1405 | 77.3821 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.4947 | 77.1374 | ursus_americanus | ENSUAMP00000035161 | 85.6593 | 77.2859 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 76.1768 | 63.5927 | monodelphis_domestica | ENSMODP00000021626 | 75.6679 | 63.1679 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 83.2853 | 73.6792 | bison_bison_bison | ENSBBBP00000002580 | 83.1256 | 73.5379 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 84.2459 | 75.9846 | monodon_monoceros | ENSMMNP00015021431 | 84.7343 | 76.4251 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 83.0932 | 73.487 | sus_scrofa | ENSSSCP00000062586 | 81.5269 | 72.1018 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 87.4159 | 79.1547 | microcebus_murinus | ENSMICP00000013168 | 87.7531 | 79.46 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 84.2459 | 75.5043 | jaculus_jaculus | ENSJJAP00000002211 | 85.063 | 76.2367 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 83.5735 | 73.8713 | ovis_aries_rambouillet | ENSOARP00020032468 | 80.9302 | 71.5349 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.4947 | 77.0413 | ursus_maritimus | ENSUMAP00000006806 | 85.6593 | 77.1896 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 84.342 | 76.2728 | delphinapterus_leucas | ENSDLEP00000027604 | 84.8309 | 76.715 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 83.3814 | 75.9846 | physeter_catodon | ENSPCTP00005021117 | 83.8647 | 76.4251 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 81.8444 | 70.7012 | mus_caroli | MGP_CAROLIEiJ_P0093387 | 82.5581 | 71.3178 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 82.1326 | 70.7012 | mus_musculus | ENSMUSP00000107793 | 82.8488 | 71.3178 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 84.9184 | 75.8886 | mustela_putorius_furo | ENSMPUP00000002538 | 81.3247 | 72.6771 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 96.2536 | 92.8914 | aotus_nancymaae | ENSANAP00000025430 | 95.702 | 92.3591 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 95.389 | 91.9308 | saimiri_boliviensis_boliviensis | ENSSBOP00000037869 | 93.7677 | 90.3683 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 84.438 | 76.4649 | vicugna_pacos | ENSVPAP00000000303 | 83.874 | 75.9542 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 97.8867 | 96.6378 | chlorocebus_sabaeus | ENSCSAP00000019029 | 97.9808 | 96.7308 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 70.8934 | 65.2257 | tupaia_belangeri | ENSTBEP00000009579 | 70.3527 | 64.7283 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 81.9404 | 71.0855 | mesocricetus_auratus | ENSMAUP00000003854 | 82.9767 | 71.9844 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 84.1499 | 75.9846 | phocoena_sinus | ENSPSNP00000021767 | 84.6377 | 76.4251 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 84.7262 | 76.9452 | camelus_dromedarius | ENSCDRP00005014619 | 81.8182 | 74.3043 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 84.7262 | 76.1768 | panthera_tigris_altaica | ENSPTIP00000018799 | 83.6019 | 75.1659 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 81.7483 | 70.5091 | rattus_norvegicus | ENSRNOP00000066040 | 82.7017 | 71.3314 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.4947 | 77.7137 | prolemur_simus | ENSPSMP00000037419 | 87.7712 | 79.783 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 80.8838 | 69.7406 | microtus_ochrogaster | ENSMOCP00000012867 | 81.5891 | 70.3488 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 81.8444 | 71.3737 | peromyscus_maniculatus_bairdii | ENSPEMP00000001641 | 82.6382 | 72.066 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 83.6695 | 73.9673 | bos_mutus | ENSBMUP00000019544 | 84.3175 | 74.5402 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 81.9404 | 70.1249 | mus_spicilegus | ENSMSIP00000025148 | 82.655 | 70.7364 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 81.7483 | 70.7012 | cricetulus_griseus_chok1gshd | ENSCGRP00001004191 | 82.8627 | 71.665 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 60.903 | 54.3708 | pteropus_vampyrus | ENSPVAP00000008908 | 60.0379 | 53.5985 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 79.5389 | 69.7406 | myotis_lucifugus | ENSMLUP00000011602 | 79.5389 | 69.7406 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 76.561 | 65.0336 | vombatus_ursinus | ENSVURP00010029270 | 76.561 | 65.0336 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 80.4995 | 70.4131 | rhinolophus_ferrumequinum | ENSRFEP00010025107 | 80.6545 | 70.5486 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 84.7262 | 75.9846 | neovison_vison | ENSNVIP00000025540 | 84.8893 | 76.1309 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 80.5956 | 71.4697 | heterocephalus_glaber_female | ENSHGLP00000001702 | 81.9336 | 72.6562 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 95.0048 | 93.3718 | rhinopithecus_bieti | ENSRBIP00000024023 | 97.3425 | 95.6693 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.6868 | 77.3295 | canis_lupus_dingo | ENSCAFP00020003260 | 85.9345 | 77.553 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.879 | 76.8492 | sciurus_vulgaris | ENSSVLP00005027110 | 84.7393 | 75.8294 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 77.3295 | 66.0903 | phascolarctos_cinereus | ENSPCIP00000040753 | 77.3295 | 66.0903 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.2065 | 75.6004 | nannospalax_galili | ENSNGAP00000023191 | 85.7834 | 76.1122 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 84.8223 | 75.7925 | chinchilla_lanigera | ENSCLAP00000025329 | 85.314 | 76.2319 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 83.6695 | 73.9673 | cervus_hanglu_yarkandensis | ENSCHYP00000035911 | 84.3175 | 74.5402 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 83.4774 | 74.0634 | catagonus_wagneri | ENSCWAP00000023758 | 84.2054 | 74.7093 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.879 | 76.7531 | ictidomys_tridecemlineatus | ENSSTOP00000005436 | 86.0443 | 76.9009 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.5908 | 74.928 | otolemur_garnettii | ENSOGAP00000021714 | 85.5908 | 74.928 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 95.9654 | 92.123 | cebus_imitator | ENSCCAP00000012451 | 94.3343 | 90.5571 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 85.7829 | 76.561 | urocitellus_parryii | ENSUPAP00010002825 | 86.2802 | 77.0048 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 83.4774 | 73.6792 | moschus_moschiferus | ENSMMSP00000015746 | 84.1239 | 74.2498 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 97.7906 | 96.1575 | rhinopithecus_roxellana | ENSRROP00000011677 | 97.7906 | 96.1575 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 82.3247 | 70.1249 | mus_pahari | MGP_PahariEiJ_P0090988 | 83.0426 | 70.7364 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 87.6081 | 80.0192 | propithecus_coquereli | ENSPCOP00000015442 | 87.7767 | 80.1732 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 65.61 | 48.415 | latimeria_chalumnae | ENSLACP00000018907 | 67.2244 | 49.6063 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 62.1518 | 46.3016 | lepisosteus_oculatus | ENSLOCP00000012206 | 62.2714 | 46.3908 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 62.8242 | 45.9174 | lepisosteus_oculatus | ENSLOCP00000012217 | 63.6187 | 46.4981 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 46.0134 | 28.9145 | clupea_harengus | ENSCHAP00000020270 | 46.1464 | 28.9981 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 44.9568 | 29.8751 | carassius_auratus | ENSCARP00000032197 | 46.1994 | 30.7009 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 47.4544 | 31.7964 | astyanax_mexicanus | ENSAMXP00000026319 | 47.6834 | 31.9498 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 46.7819 | 32.0845 | esox_lucius | ENSELUP00000028602 | 47.2816 | 32.4272 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 63.3045 | 44.3804 | esox_lucius | ENSELUP00000037072 | 63.9806 | 44.8544 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 46.6859 | 31.1239 | electrophorus_electricus | ENSEEEP00000004229 | 47.5538 | 31.7025 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 46.7819 | 32.4688 | oncorhynchus_kisutch | ENSOKIP00005099058 | 47.0077 | 32.6255 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 62.6321 | 43.1316 | oncorhynchus_kisutch | ENSOKIP00005017577 | 62.9344 | 43.3398 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 62.2478 | 43.1316 | oncorhynchus_kisutch | ENSOKIP00005041868 | 59.8338 | 41.4589 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 47.1662 | 31.9885 | oncorhynchus_kisutch | ENSOKIP00005069355 | 47.5315 | 32.2362 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 47.0701 | 32.4688 | oncorhynchus_mykiss | ENSOMYP00000059575 | 47.2973 | 32.6255 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 47.3583 | 32.3727 | oncorhynchus_mykiss | ENSOMYP00000008668 | 47.6329 | 32.5604 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 61.4793 | 42.7474 | oncorhynchus_mykiss | ENSOMYP00000060924 | 62.8684 | 43.7132 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 62.44 | 43.5159 | oncorhynchus_mykiss | ENSOMYP00000106449 | 62.8627 | 43.8104 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 61.9597 | 42.5552 | salmo_salar | ENSSSAP00000035750 | 62.3188 | 42.8019 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 63.1124 | 43.9962 | salmo_salar | ENSSSAP00000105997 | 63.4783 | 44.2512 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 46.2056 | 32.1806 | salmo_salar | ENSSSAP00000057976 | 46.4286 | 32.3359 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 46.5898 | 31.7964 | salmo_salar | ENSSSAP00000100464 | 46.9052 | 32.0116 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 45.9174 | 31.9885 | salmo_trutta | ENSSTUP00000009753 | 45.6979 | 31.8356 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 63.4006 | 44.2843 | salmo_trutta | ENSSTUP00000018677 | 63.7681 | 44.5411 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 46.6859 | 31.7964 | salmo_trutta | ENSSTUP00000097578 | 46.9112 | 31.9498 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 61.5754 | 42.5552 | salmo_trutta | ENSSTUP00000031982 | 61.9324 | 42.8019 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 13.1604 | 9.60615 | gadus_morhua | ENSGMOP00000041029 | 62.5571 | 45.6621 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 56.292 | 40.2498 | gadus_morhua | ENSGMOP00000024557 | 59.735 | 42.7115 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 57.1566 | 40.7301 | gadus_morhua | ENSGMOP00000068438 | 58.1055 | 41.4062 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 57.6369 | 40.3458 | gadus_morhua | ENSGMOP00000012342 | 58.1395 | 40.6977 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 57.1566 | 39.8655 | gadus_morhua | ENSGMOP00000036343 | 58.1623 | 40.567 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 56.7723 | 39.9616 | gadus_morhua | ENSGMOP00000045250 | 58.1121 | 40.9046 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 57.9251 | 42.171 | fundulus_heteroclitus | ENSFHEP00000033851 | 58.3172 | 42.4565 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 57.3487 | 41.4025 | fundulus_heteroclitus | ENSFHEP00000015612 | 59.4622 | 42.9283 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.2699 | 42.3631 | xiphophorus_maculatus | ENSXMAP00000004435 | 60.1365 | 42.9825 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 58.7896 | 42.3631 | oryzias_latipes | ENSORLP00000022276 | 58.9028 | 42.4447 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.2699 | 41.4986 | cyclopterus_lumpus | ENSCLMP00005032121 | 59.9611 | 41.9825 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 58.1172 | 41.5946 | oreochromis_niloticus | ENSONIP00000074844 | 57.4004 | 41.0816 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.366 | 42.4592 | oreochromis_niloticus | ENSONIP00000068121 | 59.4231 | 42.5 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 60.1345 | 43.4198 | oreochromis_niloticus | ENSONIP00000066484 | 60.3664 | 43.5873 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 12.1998 | 8.64553 | haplochromis_burtoni | ENSHBUP00000020363 | 50.1976 | 35.5731 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 15.562 | 11.2392 | haplochromis_burtoni | ENSHBUP00000013509 | 62.3077 | 45 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 58.3093 | 42.171 | astatotilapia_calliptera | ENSACLP00000009763 | 58.6473 | 42.4155 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.366 | 43.5159 | astatotilapia_calliptera | ENSACLP00000009787 | 59.7101 | 43.7681 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 55.6196 | 40.7301 | astatotilapia_calliptera | ENSACLP00000009783 | 59.2025 | 43.3538 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.0778 | 42.8434 | sparus_aurata | ENSSAUP00010036840 | 61.5 | 44.6 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.2699 | 43.0355 | lates_calcarifer | ENSLCAP00010013343 | 59.4986 | 43.2015 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 67.0509 | 51.3929 | xenopus_tropicalis | ENSXETP00000077686 | 67.0509 | 51.3929 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 67.0509 | 50.3362 | xenopus_tropicalis | ENSXETP00000062585 | 67.3095 | 50.5304 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 71.6619 | 57.0605 | chrysemys_picta_bellii | ENSCPBP00000017258 | 72.4272 | 57.6699 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.6542 | 42.2671 | chrysemys_picta_bellii | ENSCPBP00000017262 | 62.664 | 44.3996 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 64.073 | 48.8953 | sphenodon_punctatus | ENSSPUP00000004305 | 64.8833 | 49.5136 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 60.2305 | 48.0307 | sphenodon_punctatus | ENSSPUP00000004314 | 72.7378 | 58.0046 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 64.1691 | 47.4544 | crocodylus_porosus | ENSCPRP00005008859 | 64.2308 | 47.5 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 71.3737 | 56.3881 | crocodylus_porosus | ENSCPRP00005008857 | 69.8308 | 55.1692 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 14.6013 | 9.51009 | neolamprologus_brichardi | ENSNBRP00000011280 | 58.6873 | 38.2239 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 17.9635 | 12.1998 | neolamprologus_brichardi | ENSNBRP00000011258 | 54.3605 | 36.9186 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 63.2085 | 44.6686 | erpetoichthys_calabaricus | ENSECRP00000017341 | 63.8215 | 45.1018 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 60.8069 | 44.4765 | erpetoichthys_calabaricus | ENSECRP00000017324 | 62.6733 | 45.8416 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 57.9251 | 42.4592 | kryptolebias_marmoratus | ENSKMAP00000023702 | 58.1485 | 42.623 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.5581 | 43.1316 | oryzias_javanicus | ENSOJAP00000026696 | 60.3113 | 43.677 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.8463 | 42.5552 | amphilophus_citrinellus | ENSACIP00000010819 | 60.1351 | 42.7606 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.0778 | 42.4592 | amphiprion_percula | ENSAPEP00000033922 | 59.9415 | 43.0799 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 58.2133 | 42.4592 | oryzias_sinensis | ENSOSIP00000025943 | 60.7823 | 44.333 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 27.8578 | 20.9414 | pelodiscus_sinensis | ENSPSIP00000000427 | 68.2353 | 51.2941 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 28.9145 | 22.2863 | pelodiscus_sinensis | ENSPSIP00000000439 | 67.1875 | 51.7857 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 70.6052 | 57.0605 | pelodiscus_sinensis | ENSPSIP00000011220 | 70.8775 | 57.2806 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 66.8588 | 49.952 | gopherus_evgoodei | ENSGEVP00005008868 | 67.052 | 50.0963 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 48.6071 | 37.1758 | gopherus_evgoodei | ENSGEVP00005008924 | 68.8435 | 52.6531 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 70.7012 | 56.3881 | gopherus_evgoodei | ENSGEVP00005009041 | 69.763 | 55.6398 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 70.7973 | 56.7723 | chelonoidis_abingdonii | ENSCABP00000010648 | 70.1905 | 56.2857 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 66.3785 | 49.1835 | chelonoidis_abingdonii | ENSCABP00000015447 | 66.8279 | 49.5164 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.8463 | 43.4198 | seriola_dumerili | ENSSDUP00000004379 | 60.4854 | 43.8835 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 55.9078 | 40.5379 | cottoperca_gobio | ENSCGOP00000050213 | 58.9069 | 42.7126 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 58.0211 | 41.5946 | gasterosteus_aculeatus | ENSGACP00000005236 | 59.2738 | 42.4926 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 52.5456 | 36.5994 | cynoglossus_semilaevis | ENSCSEP00000015655 | 55.4767 | 38.641 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.2699 | 42.7474 | poecilia_formosa | ENSPFOP00000027487 | 60.1365 | 43.3723 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.9424 | 43.1316 | sander_lucioperca | ENSSLUP00000000603 | 60.9375 | 43.8477 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.1739 | 42.5552 | amphiprion_ocellaris | ENSAOCP00000012904 | 60.039 | 43.1774 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 71.7579 | 57.5408 | terrapene_carolina_triunguis | ENSTMTP00000009815 | 71.9653 | 57.7071 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 66.8588 | 49.0874 | terrapene_carolina_triunguis | ENSTMTP00000003334 | 66.6029 | 48.8995 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 71.5658 | 57.4448 | terrapene_carolina_triunguis | ENSTMTP00000003344 | 71.7726 | 57.6108 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 66.8588 | 49.0874 | terrapene_carolina_triunguis | ENSTMTP00000019058 | 66.6029 | 48.8995 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 63.1124 | 43.2277 | hucho_hucho | ENSHHUP00000041034 | 63.417 | 43.4363 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 46.9741 | 31.4121 | hucho_hucho | ENSHHUP00000004872 | 47.3837 | 31.686 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 63.0163 | 43.9962 | hucho_hucho | ENSHHUP00000011672 | 63.4429 | 44.294 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 46.0134 | 32.0845 | hucho_hucho | ENSHHUP00000036932 | 46.55 | 32.4587 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 46.1095 | 30.8357 | pygocentrus_nattereri | ENSPNAP00000024411 | 46.9208 | 31.3783 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 58.9817 | 42.171 | labrus_bergylta | ENSLBEP00000026945 | 60.0783 | 42.955 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 33.9097 | 24.7839 | pundamilia_nyererei | ENSPNYP00000015337 | 34.4055 | 25.1462 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 58.1172 | 41.8828 | pundamilia_nyererei | ENSPNYP00000015358 | 58.3977 | 42.0849 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 60.2305 | 42.8434 | mastacembelus_armatus | ENSMAMP00000019936 | 60.8738 | 43.301 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 58.8857 | 41.8828 | stegastes_partitus | ENSSPAP00000010486 | 59.6884 | 42.4538 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 62.9203 | 43.5159 | oncorhynchus_tshawytscha | ENSOTSP00005068429 | 63.2239 | 43.7259 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 46.9741 | 31.9885 | oncorhynchus_tshawytscha | ENSOTSP00005044807 | 47.3379 | 32.2362 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 46.9741 | 32.1806 | oncorhynchus_tshawytscha | ENSOTSP00005032338 | 47.2008 | 32.3359 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 62.9203 | 43.5159 | oncorhynchus_tshawytscha | ENSOTSP00005060934 | 63.2239 | 43.7259 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 62.3439 | 43.5159 | oncorhynchus_tshawytscha | ENSOTSP00005040025 | 59.9261 | 41.8283 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.1739 | 42.3631 | acanthochromis_polyacanthus | ENSAPOP00000018638 | 60.039 | 42.9825 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 54.5629 | 39.3852 | nothobranchius_furzeri | ENSNFUP00015019198 | 58.1372 | 41.9652 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 58.5975 | 43.708 | cyprinodon_variegatus | ENSCVAP00000011182 | 32.4296 | 24.1893 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 60.903 | 42.3631 | denticeps_clupeoides | ENSDCDP00000034422 | 61.079 | 42.4855 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.0778 | 41.8828 | larimichthys_crocea | ENSLCRP00005025936 | 58.2386 | 41.2879 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 60.0384 | 43.2277 | hippocampus_comes | ENSHCOP00000019085 | 61.2145 | 44.0744 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 45.1489 | 30.2594 | cyprinus_carpio_carpio | ENSCCRP00000103503 | 46.4427 | 31.1265 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.366 | 42.9395 | poecilia_latipinna | ENSPLAP00000031085 | 59.8257 | 43.272 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 45.0528 | 30.3554 | sinocyclocheilus_grahami | ENSSGRP00000035431 | 46.2525 | 31.1637 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 60.8069 | 42.6513 | paramormyrops_kingsleyae | ENSPKIP00000031136 | 61.8768 | 43.4018 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 35.3506 | 23.1508 | scleropages_formosus | ENSSFOP00015007176 | 47.5452 | 31.137 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 65.8982 | 49.3756 | leptobrachium_leishanense | ENSLLEP00000005747 | 66.3443 | 49.7099 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 66.6667 | 49.0874 | leptobrachium_leishanense | ENSLLEP00000005732 | 66.7308 | 49.1346 |
| ENSG00000101916 | homo_sapiens | ENSP00000218032 | 59.2699 | 43.0355 | oryzias_melastigma | ENSOMEP00000005753 | 60.0195 | 43.5798 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000139 | located_in Golgi membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0001932 | involved_in regulation of protein phosphorylation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0002224 | involved_in toll-like receptor signaling pathway | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0003677 | enables DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0003725 | enables double-stranded RNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003727 | enables single-stranded RNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005789 | located_in endoplasmic reticulum membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005886 | is_active_in plasma membrane | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0006954 | involved_in inflammatory response | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007249 | involved_in I-kappaB kinase/NF-kappaB signaling | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0009615 | involved_in response to virus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0009897 | located_in external side of plasma membrane | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0010008 | located_in endosome membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0016064 | involved_in immunoglobulin mediated immune response | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0032695 | involved_in negative regulation of interleukin-12 production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032727 | involved_in positive regulation of interferon-alpha production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032728 | involved_in positive regulation of interferon-beta production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032729 | involved_in positive regulation of type II interferon production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032731 | involved_in positive regulation of interleukin-1 beta production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032755 | involved_in positive regulation of interleukin-6 production | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0032757 | involved_in positive regulation of interleukin-8 production | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0034158 | involved_in toll-like receptor 8 signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0036020 | located_in endolysosome membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0038023 | enables signaling receptor activity | 1 | NAS | Homo_sapiens(9606) | Function |
| GO:0038187 | enables pattern recognition receptor activity | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0045087 | involved_in innate immune response | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0045089 | involved_in positive regulation of innate immune response | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051607 | involved_in defense response to virus | 4 | IBA,IDA,IEP,IMP | Homo_sapiens(9606) | Process |
| GO:0071260 | involved_in cellular response to mechanical stimulus | 1 | IEP | Homo_sapiens(9606) | Process |